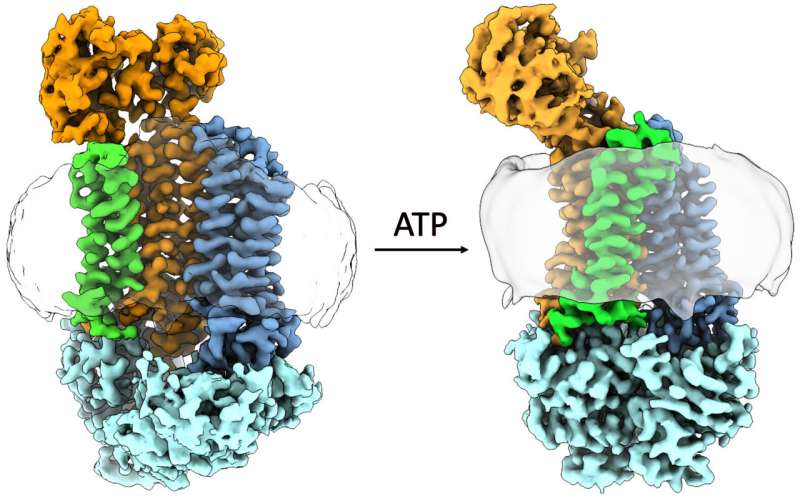
Researchers have shown how a protein implicated in antibiotic resistance changes it conformation when given ATP, often called the fuel of cells. Credit: Orlando Lab/PNAS
Michigan State University's Ben Orlando is a structural biologist who studies some of nature's smallest machines, sees how they are put together and figures out how they work. He's currently focused on proteins that bacteria use to survive antibiotic treatments so that he can help decommission these biological machines and fight potentially deadly infections.
Orlando's team has now taken new, atomically detailed snapshots of a bacterial protein that helps many germs sense and evade antibiotics. The researchers published their work March 29 in the journal Proceedings of the National Academy of Sciences.
"What our research does is help set the groundwork to understand how a variety of human pathogens can resist some of our most powerful medicines," said Orlando, an assistant professor in the College of Natural Science.
Antibiotic resistance is one of the biggest threats to global health, according to the World Health Organization. In the United States alone, antibiotic resistant bacteria are responsible for nearly 3 million infections and 35,000 deaths each year, per the Centers for Disease Control and Prevention.
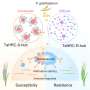
For their work, Orlando and his team are focused on a protein that contributes to antibiotic resistance by protecting certain bacteria from what are known as antimicrobial peptides. These molecules include the bacitracin found in Neosporin and other topical ointments as well as vancomycin, which is used to treat a range of tough infections.
The researchers zoomed in on a protein that lives on the outer membrane of many Gram-positive bacteria, where it acts as a sentry to guard the cell from antibiotic attack. Evolution has crafted the protein machinery to recognize peptides, latch onto them and then flick them away before they can harm the cell.
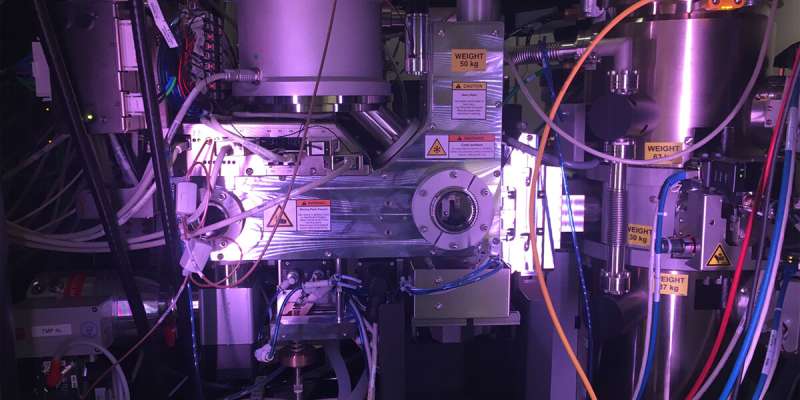
A look inside MSU’s cryogenic electron microscope. Credit: Courtesy of Ben Orlando
"If we can understand how these machines work, then maybe we can jam them up with new molecules or develop new antibiotics that evade resistance," said Orlando, who was hired in 2020 as part of MSU's Global Impact Initiative.
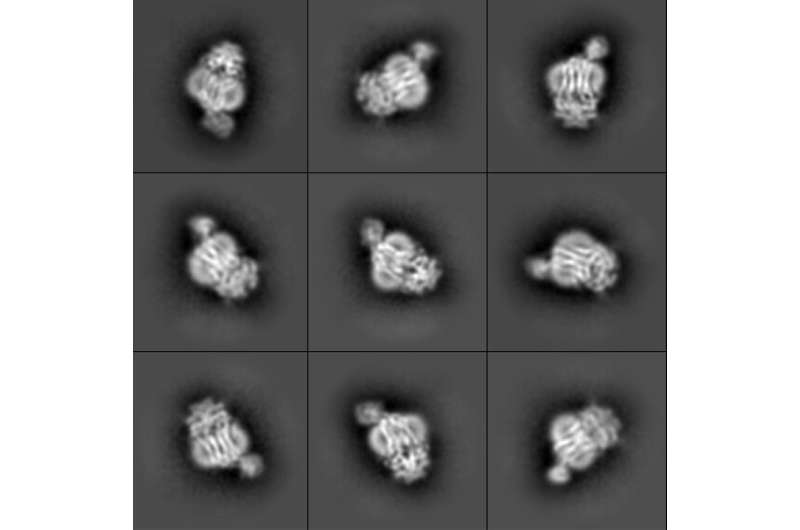
The research team used a technique known as cryogenic electron microscopy, or cryo-EM, to image the bacterial protein complex. First, the team created pure samples of the nanosized proteins they were studying. Those proteins then went into a water-based solution that the researchers used to coat a sample holder, or grid. The researchers then dip the grid in liquid ethane that freezes the water so fast it can't crystallize, resulting in a glass-like ice.
Using the electron microscope in MSU's Cryo-EM Facility, the researchers took thousands of snapshots of the protein in different orientations. From those images, the researchers built up the 3D atomic structure of the biological machine and saw how it changed its conformation with help from ATP, or adenosine triphosphate, a molecule that fuels biochemical processes.
"This structure is quite unlike anything we've seen before," Orlando said. "My favorite part of the job is when that data comes in, we can build that atomic model and see the overall structure before anyone else."
"These proteins are very difficult to study given their size and flexibility," said Kristin N. Parent, director of the Cryo-EM Facility and J.K. Billman Jr., M.D., Endowed Research Professor. "Dr. Orlando is providing a crucial glimpse into how these complexes work."
More than 30 groups at MSU—and many from outside of Michigan State—rely on the tools and know-how at MSU's Cryo-EM Facility, which is part of MSU's Research Technology Support Facility, or RTSF. The RTSF is also home to the Mass Spectrometry and Metabolomics Core that provided key support and expertise for the project, which was spearheaded by Natasha George, a doctoral student in Orlando's lab.
A sampling of the cryo-EM snapshots that researchers took show the protein in different orientations. Credit: Orlando Lab/PNAS
The collaborative nature of the project makes it that much more rewarding, especially in the rapidly developing field of cryo-EM, Orlando said. When he was a postdoctoral researcher at Harvard Medical School, he said he would have to take those thousands of electron micrograph snapshots by hand.
Now, processes are much more automated, accelerating the timeline for discoveries when you have the right team.
"This protein is on the harder end of the spectrum as far as samples go—we are continually looking to push the limits of what's possible with this technology," Orlando said. "Natasha George put a lot of effort into this and made it come to fruition. I'm really proud of the work she's done!"
Which underscores another advantage of doing this type of cutting-edge work in a growing scientific field on the banks of the Red Cedar.
"Not only are we doing cool science, but, here at MSU, we're training the next generation of scientists who will carry on that work," Orlando said.
More information: Natasha L. George et al, Conformational snapshots of the bacitracin sensing and resistance transporter BceAB, Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2123268119
Journal information: Proceedings of the National Academy of Sciences
Provided by Michigan State University