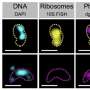
Research groups from the University of Groningen have revealed a novel mechanism of ribosome dimerization in the bacterium Lactococcus lactis using cryo-electron microscopy. As this dimerization renders ribosomes more resistant to antibiotics, this study provides the necessary structural basis to design new generations of antibiotics. The results are published in Nature Communications on Sept. 28.
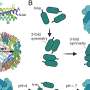
Antibiotics are the most common medication used to treat microbial infections. Many antibiotics target intracellular bacterial ribosomes—cellular factories that synthesize proteins—which are essential for bacterial survival and proliferation. When bacteria have an excess of protein synthesis activity, they stall the ribosomes in an inactive dimeric complex (i.e. two copies of ribosomes interact with each other). This so-called hibernating ribosome complex is more resistant to antibiotics.
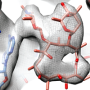
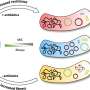
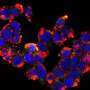
In a collaborative effort, research groups led by Egbert Boekema, Bert Poolman and Albert Guskov revealed a novel mechanism of ribosome dimerization in the bacterium Lactococcus lactis using cryo-electron microscopy. The peculiarity of the mechanism they describe is that it involves a single protein, named HPFlong, which is able to dimerize on its own and then pull two copies of ribosomes together. The dimeric state of the ribosome is no longer capable of synthesizing new proteins.
This hibernation mechanism is in a stark contrast with previous studies done in another microorganism, Escherichia coli. However, based on a phylogenetic analysis of the amino acid sequence of HPFlong, the researchers conclude that the mechanism they propose is more widespread, since protein HPFlong is present in nearly all known bacteria. This study provides the necessary structural basis to design new generations of antibiotics targeting hibernating ribosomes.
More information: Linda E. Franken et al, A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy, Nature Communications (2017). DOI: 10.1038/s41467-017-00718-x
Journal information: Nature Communications
Provided by University of Groningen