Orthosomycins bind to bacterial ribosome site not used by any other antibiotic
An LMU team has shown that orthosomycins bind to a site on the bacterial ribosome that is not used by any other antibiotic. The finding could lead to agents that are effective against existing multi-drug resistant bacterial strains.
Bacterial pathogens that have become resistant to several different classes of antibiotics, and for which no safe and effective alternative therapies are currently available, are a steadily growing problem. Hence there is already a pressing need for new antibacterial drugs. Biochemical investigations have suggested that compounds known as orthosomycins could provide the basis for the design of antibiotics with novel modes of action. Now, researchers led by Dr. Daniel Wilson at the LMU Gene Center, in collaboration with teams led by Dr. Scott Blanchard at Cornell University (Ithaca, New York) and Dr. Yury Polikanov of the University of Illinois in Chicago, have characterized the complexes formed by two orthosomycins with their target, and provided additional insights into their mechanism of action. The new findings, which appear in the latest issue of the journal PNAS, provide the basis for further optimization of these agents for use against multi-drug resistant bacterial strains.
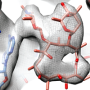
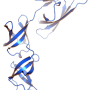
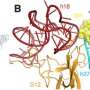
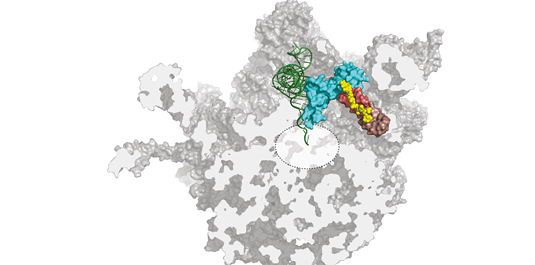
Orthosomycins are unusual in having a relatively extended structure but, like many other antibiotics, they bind to bacterial ribosomes. Ribosomes are the subcellular organelles responsible for protein synthesis, both in bacteria and higher organisms. Because the ribosomes found in the nucleated cells of the latter differ in some respects from their bacterial counterparts, antibiotics can inhibit the synthesis of bacterial proteins without affecting the production of host proteins. "With the aid of high-resolution cryo-electron micrographs, we have identified the binding sites for two orthosomycins, evernimicin and avilamycin, on the bacterial ribosome," says Stefan Arenz, a member of Wilson's research group and first author of the new study. "And the results reveal that they both interact with a site that is not recognized by any other class of antibiotic."
Bacterial ribosomes are made up of three ribosomal RNA (rRNA) molecules, which are folded into complex shapes and serve as scaffolds for the assembly of more than 50 different proteins. Evernimicin and avilamycin bind in the groove between two short double-stranded stretches in one of the rRNAs, such that they also interact with a specific ribosomal protein named L16. Unlike mutations in regions with clustered binding sites, "mutations here cannot lead to cross-resistance to any other sort of antibiotic, because this site is utilized only by orthosomycins," Arenz points out. Further analysis of the effect of binding on ribosomal function, using the Fluorescence Resonance Energy Transfer (FRET) technique, enabled the researchers to define how the orthosomycins inhibit the elongation phase of protein synthesis.
The blueprint for the synthesis of any given protein is encoded in the nucleotide sequence of a messenger RNA (mRNA). The ribosome works basically by sequentially matching trinucleotide sequences in the mRNA with nucleotide triplets on so-called transfer RNAs (tRNAs) to which the appropriate amino acid is attached. The overall result is that a defined nucleotide sequence is translated into a particular sequence of amino acids, i.e. a protein with a specific function. During this process, one end of each tRNA must first bind to the ribosome so that the other end can swivel into position in the organelle's active center, to allow the new amino acid to be linked to the preceding one. "The evernimicin/avilamycin binding site partially overlaps the region that would normally accommodate the hinge segment of the tRNA," Arenz explains. "So prior binding of either antibiotic prevents the swiveling movement, and brings the ribosome to a standstill."
More information: Stefan Arenz et al. Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome, Proceedings of the National Academy of Sciences (2016). DOI: 10.1073/pnas.1604790113
Journal information: Proceedings of the National Academy of Sciences
Provided by Ludwig Maximilian University of Munich