This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Innovative microscopy reveals amyloid architecture, may give insights into neurodegenerative disease
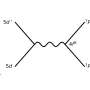
Amyloid-beta (A-beta) aggregates are tangles of proteins most notably associated with neurodegenerative diseases such as Alzheimer's. Despite its constant stint in the limelight, however, researchers have been unable to get a good understanding of how A-beta comes together and breaks apart.
"The way A-beta behaves in a variety of environments, including the human brain, is elusive," said Brian Sun, an electrical systems and engineering alumnus of Washington University in St. Louis who is now an MD/Ph.D. student at the School of Medicine. "There's an understanding of growth and decay that isn't fully fleshed out."
That's going to change, thanks to research recently published in Nano Letters by Sun with colleagues in Matthew Lew's lab in the Preston M. Green Department of Electrical & Systems Engineering at WashU's McKelvey School of Engineering.
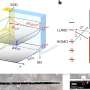
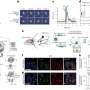
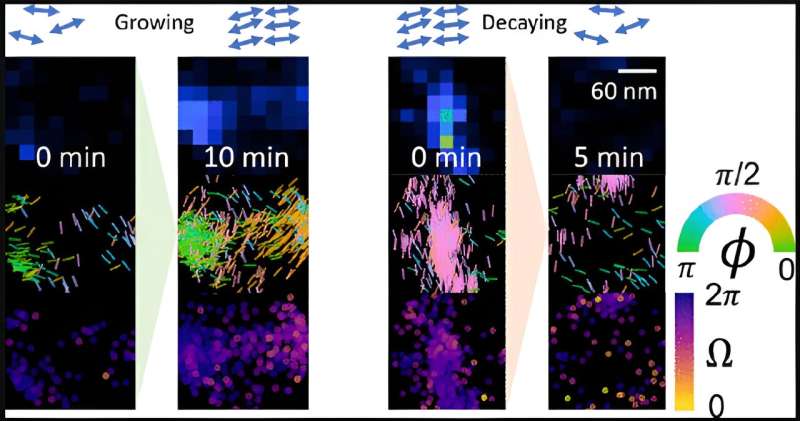
In first-of-its-kind work, Sun and colleagues were able to measure amyloid fibril beta-sheet assemblies, the underlying girders of the protein conglomeration, while they were changing. Previous high-resolution microscopy studies have only gotten static shots.
"We wanted to look specifically at dynamics of the underlying structure of A-beta that could be responsible for the changes we're seeing, not just changes in the overall shape," said Sun, the paper's first author.
Lew used Lego bricks in an analogy, noting that current imaging technology shows you the full Lego building but not a look at how individual bricks are organized.
"The individual proteins are always changing in response to their environment," said Associate Professor Lew. "It is like having certain Lego bricks causing other bricks to change their shape. The changing architecture of the proteins and the assembled aggregates together leads to the complexity of neurodegenerative disease."
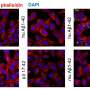
The Lew lab has developed a new type of imaging tech that allows researchers to see the orientation and other minute details in nanostructures of biological systems that were previously invisible. Their technique—single-molecule orientation–localization microscopy (SMOLM)—uses flashes of light from chemical probes to visualize the sheets of peptides underlying Aβ42, one kind of A-beta peptide.
Using SMOLM lets them look at individual orientation of the underlying beta-sheets to see the relationship between their organization and how that relates to the amyloid protein's overall structure.
Multiple ways to remodel
Aβ42 is constantly changing, and step one is to try to find a method to the madness, a model or pattern of action to predict the protein's behavior.
Now that the Lew lab can make these measurements, they made some intuitive observations and found some surprises hidden in the amyloid-beta architecture.
As expected, stable Aβ42 structures tend to retain stable underlying beta-sheets; growing structures have underlying beta-sheets that become more defined and rigid as the growth continues. Decaying structures exhibit increasingly disordered and less rigid beta-sheets. But they also found more than one way that Aβ42 can renovate.
"There are multiple different ways for Aβ42 structures to remain stable, or grow and decay," Sun said.
The researchers also discovered that Aβ42 can grow and decay in ways that defy expectations. For example, Aβ42 can grow and decay in ways that preserve the underlying structure; sometimes there's growth where the peptides just pile on, but the underlying beta-sheet orientations don't change. In other cases, Aβ42 undergoes "stable decay," where the opposite happens, i.e., peptides leave, but beta-sheet structure remains.
Finally, Aβ42's beta-sheets sometimes reorganize and change orientations without immediate accompanying changes to the overall shape. These nano-structural reorganizations can predispose to future large-scale remodeling.
"Because SMOLM can track Aβ42's underlying organization and not just its shape, we can see different kinds of subtypes of remodeling that aren't visible to diffraction-limited, non-orientation imaging modalities," Sun said.
If it all sounds a bit vague, keep in mind this is the first pass at even looking at these constantly shifting nanoscale structures. It's all the more notable that Sun crafted this work while juggling COVID-19 lockdown restrictions and his undergraduate course load at WashU, which he completed in three years. It paves the way for him and others to really get a handle on amyloid architecture.
During the graduate phase of his MD/Ph.D. training, Sun plans to design nanoscale imaging systems and sensors that could reveal the hidden mechanisms of difficult-to-treat diseases.
Sun credits McKelvey Engineering and the Lew lab for the rigorous training that made this study and academic trajectory possible, as well as the MSTP for supporting his continued research post-graduation. "I'm really glad I went through this journey," he said.
More information: Brian Sun et al, Single-Molecule Orientation Imaging Reveals the Nano-Architecture of Amyloid Fibrils Undergoing Growth and Decay, Nano Letters (2024). DOI: 10.1021/acs.nanolett.4c01263
Journal information: Nano Letters
Provided by Washington University in St. Louis