Ribosome recycling as a drug target
Ludwig Maximilian University of Munich researchers have elucidated a mechanism that recycles bacterial ribosomes stalled on messenger RNAs that lack termination codons. The protein involved provides a potential target for future antibiotics.
So-called multidrug-resistant bacteria - strains that have become resistant to conventional antibiotics - pose an increasingly serious medical problem, which underlines the urgent need to develop new antibiotics that attack novel targets. "Many of the most effective antibiotics act by inhibiting various steps in protein synthesis on bacterial ribosomes," as LMU biochemist Daniel Wilson points out. The search is now on for agents that interfere with aspects of the process that have so far been overlooked in this context. One of these processes is the recycling of stalled ribosomes. Wilson and his research group have just completed a comprehensive structural study of ribosome recycling. The results of the study, which appears in the leading journal Nature, may identify promising points of attack for future antibiotics.
Ribosomes are the organelles that translate the nucleotide sequences encoded in messenger RNA molecules (mRNAs) into the amino-acid sequences of the corresponding proteins. The ribosome "reads" the nucleotide sequence in a fixed direction and is released from the mRNA only when it reaches a defined termination signal. Errors in synthesis or processing may, however, lead to the production of mRNAs that lack termination signals, causing the ribosome to stall while still attached to the mRNA and the growing protein. Cells have evolved several ways of detaching stalled ribosomes from the truncated mRNAs and recycling them for re-use.
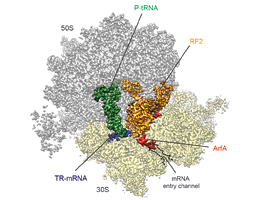
Wilson's team has now employed cryo-electron microscopy to determine the structure of the bacterial recycling factor ArfA. Their analysis reveals how ArfA recognizes the stalled ribosome on a defective mRNA and recruits a so-called release factor, which detaches the incomplete protein. This then enables the stalled ribosome to be released from the mRNA in the normal manner by dissociation into its two component subunits. The subunits are then free to interact with another mRNA, thus allowing protein synthesis to proceed.
"Our results could facilitate the development of new antibiotics that act by inhibiting ArfA-mediated ribosome recycling," says Wilson. Since recycling of human ribosomes is dependent on factors that are unrelated to ArfA, such inhibitors should act specifically on the bacterial ribosome, and kill the bacterial cell by gradually cutting off the supply of free ribosomes.
More information: Paul Huter et al, Structural basis for ArfA–RF2-mediated translation termination on mRNAs lacking stop codons, Nature (2016). DOI: 10.1038/nature20821
Journal information: Nature
Provided by Ludwig Maximilian University of Munich