Researchers discover how hibernating ribosomes wake up

In research published in Proceedings of the National Academy of Sciences (PNAS), Mee-Ngan F. Yap, Ph.D., assistant professor of biochemistry and molecular biology at Saint Louis University, has uncovered the way a bacterial ribosome moves from an inactive to an active form, and how that "wake up call" is key to its survival.
Often described as a cell's protein factory, ribosomes translate messenger RNA and link amino acids together to form new proteins. Ribosomes catalyze proteins that are essential for all life.
In bacteria, ribosomes can take an inactive form called hibernating 100S ribosome. Because protein synthesis accounts for more than half of a cell's energy costs, the inactive ribosome form helps bacteria survive under stressful conditions. During limited nutrient access, antibiotic stress, host colonization, adaptation to the dark and biofilm formation, bacteria aim to conserve energy by shutting down the protein factory.
Scientists have observed that the hibernating form of the ribosome is not a permanent state and that if conditions are favorable, it can "wake up" and return to its active form, called 70S, and begin to initiate new cycles of protein synthesis.
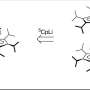
"The 100S form is not held together forever," Yap said. "However, until now, the disassociation of 100S ribosome has been a complete black box. We haven't known how ribosomes move from one form to the other."
Yap was looking for the protein factor that caused the 100S form to return to the intermediate 30S and 50S forms and subsequently into the active 70S form. Studying Staphylococcus aureus, commonly known as staph, Yap found that a GTP hydrolase enzyme called HflX is the wake-up call that will re-activate the ribosome.
"HflX is one way to break up the 100S ribosome structure so that it can return to the active 70S form," Yap said.
HflX GTPases are a family of enzymes that are evolutionarily conserved proteins, meaning they also exist in plants, humans and other bacteria. Yap is intrigued by this finding, because while there has been virtually no study of the protein in human cells, it appears in genetic sequencing mapped to cancer patients and those with neurological symptoms, including tic disorder-like syndromes. Scientists do not yet know what this connection means.
Thanks to a new five-year $1.59 million grant from the National Institutes of Health, Yap will continue to explore these questions and others that surround hibernating ribosomes' role in cell survival.
More information: Arnab Basu et al, Disassembly of the Staphylococcus aureus hibernating 100S ribosome by an evolutionarily conserved GTPase, Proceedings of the National Academy of Sciences (2017). DOI: 10.1073/pnas.1709588114
Journal information: Proceedings of the National Academy of Sciences
Provided by Saint Louis University