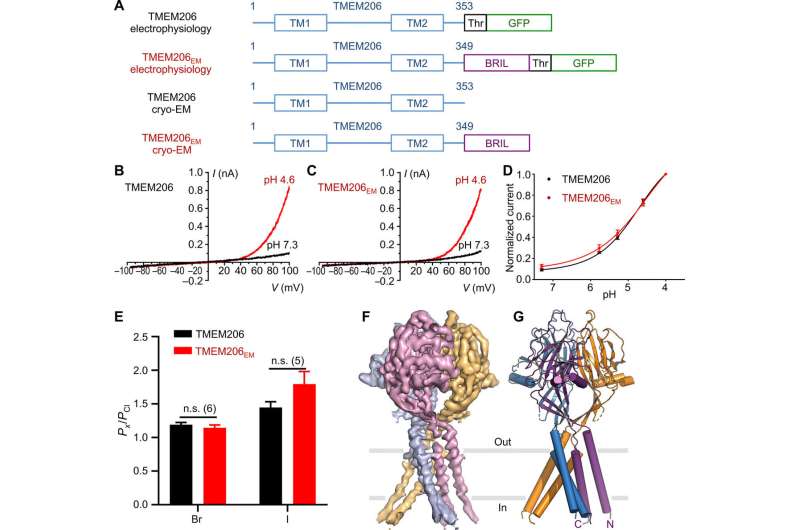
Function and structure of pufferfish TMEM206. (A) Schematic of channel constructs used for electrophysiology and single-particle cryo-EM experiments. “Thr” represents a thrombin cleavage site. (B and C) Representative whole-cell current traces activated by extracellular pH 4.6 for pufferfish TMEM206 (B) and TMEM206EM (C). Channel constructs were expressed in TMEM206 knockout human embryonic kidney (HEK) 293T cells. (D) Normalized current-to-pH relationships of pufferfish TMEM206 (n = 6 to 9 cells per data point) and TMEM206EM (n = 5 to 6 cells per data point). All currents were recorded at room temperature and normalized to pH 4.0 currents at +100 mV. (E) Anion selectivity for pufferfish TMEM206 and TMEM206EM. Data are presented as means ± SEM (n.s., not significant; Student’s t test). (F) Cryo-EM density of pufferfish TMEM206EM contoured at 7.0 σ and colored by individual subunits. (G) Trimeric structure of pufferfish TMEM206EM. Credit: Science Advances, doi: 10.1126/sciadv.abe5983
The transmembrane protein 206 abbreviated as TMEM206 is an evolutionarily conserved chloride channel that underlies the ubiquitously expressed, proton-activated, outwardly rectifying anion currents. In a new report now published on Science Advances, Zengqin Deng and a multidisciplinary research team at the Washington University School of Medicine, in Saint Louis, U.S. described the cryo-electron microscopy (cryo-EM) structure of the pufferfish TMEM206. The structure formed a trimeric channel with two transmembrane segments and a large extracellular domain. According to the results, Deng et al. showed how an ample vestibule in the extracellular region could be accessed laterally from three side portals, where the central pore contained multiple constructions. For instance, a conserved lysine residue close to the cytoplasmic region of the inner helix, presumably formed the chloride ion selectivity filter. The core structure and assembly resembled those of sodium channels that are unrelated in amino acid sequence, and therefore conduct cations instead of anions. Together with electrophysiology they provided insights on ion conduction and gating for a new class of chloride channels that are architecturally distinct from previously described chloride channel families.
Chloride ion channels
Chloride ions are abundant anions in animals, and they move across cell membranes via chloride channels and transporters for a variety of cellular functions, including cell volume regulation, intracellular acidification and excitability control in muscles. The ions are widely observed in mammalian cells although the molecular components behind the chloride currents have remained elusive until now. Two independent studies that used genome-wide RNA interference screening had identified TMEM206 as the underlying anion channel. The TMEM206 is evolutionally conserved in vertebrates. Chloride channels are diverse in both amino acid sequence and three-dimensional architecture. In this work, Deng et al. presented a cryo-electron microscopy (cryo-EM) structure of pufferfish TMEM206 to reveal a trimeric channel architecture different from those previously known. The scientists combined electrophysiology with this work to provide the first structural and functional description of an evolutionarily conserved and broadly expressed chloride channel to establish a molecular framework and understand chloride conduction and channel gating.
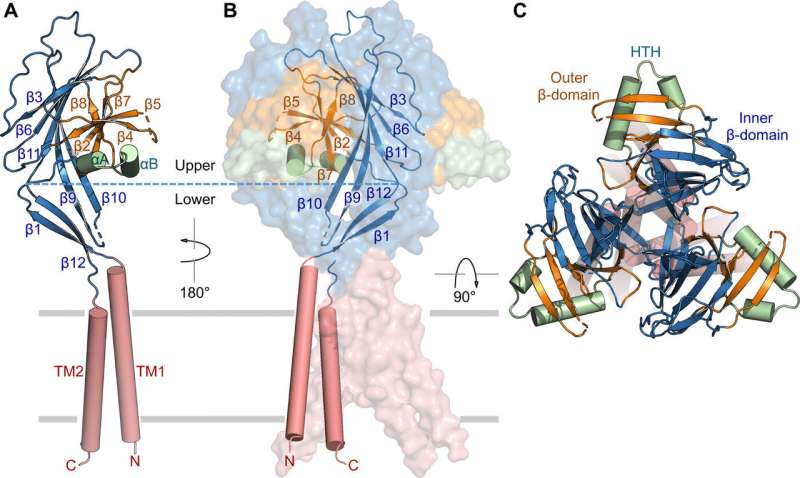
Subunit structure and channel assembly. (A) Structure of a single subunit, showing the transmembrane domain (red), inner β-domain (blue), outer β-domain (orange), and HTH (green). Secondary structure elements are indicated. (B) Trimeric channel assembly. Two of the subunits are shown in surface representation. (C) Orthogonal view as in (B), from the extracellular side. Credit: Science Advances, doi: 10.1126/sciadv.abe5983
Fusion strategy to determine the cryo-EM structure.
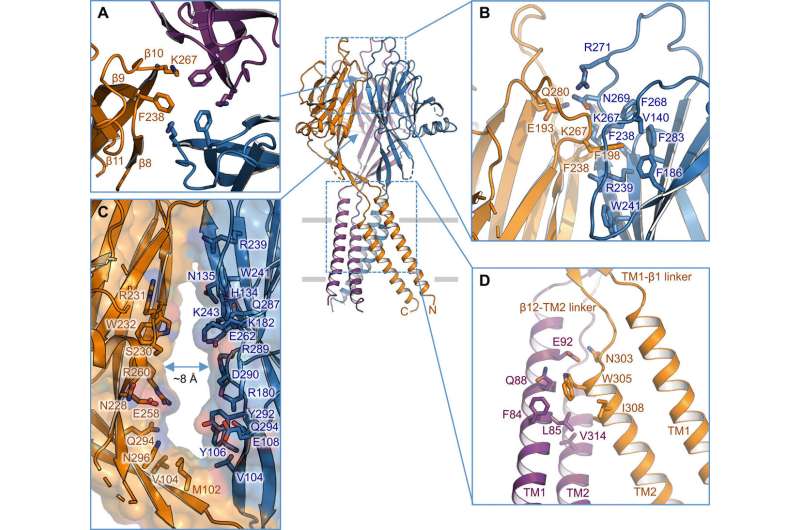
The researchers used green fluorescent protein-tagged constructs to identify TMEM206 candidates and selected pufferfish TMEM206 due to their 50 percent shared sequence identity to the human chloride channel. Deng et al. then purified the full-length wild-type pufferfish TMEM206 protein and subjected it to single-particle cryo-EM analysis. Using 3D reconstruction maps, they revealed a trimeric channel architecture with transmembrane and extramembrane domains. The TMEM206 formed a symmetric trimer, where each subunit contained a transmembrane domain (TMD) with two membrane-spanning helices named TM1 and TM2 and a large extracellular domain (ECD) enriched with β-domains with additional motifs further organized in the upper and lower layers. Additionally, extensive side-chain contacts were involved through van der Waals interactions in the inner and outer-β domains. The trimeric channel assembly introduced three lateral openings or side portals in the middle of the extracellular region to likely facilitate the passage of ion and water. The elongated side portals extended to the ECD-TMD junction for tight packing interactions to resume. To overcome any technical difficulties of determining the structure of the protein, Deng et al. fused the C-terminal of bone-restricted interferon-induced transmembrane-like protein (abbreviated BRIL); a four-helix bundle protein widely used as a crystallization chaperone to improve membrane stability and promote crystal formation.
Intersubunit interface. (A) Trimeric interface at the apex of the ECD. Side chains of K267 and F238 are highlighted. (B) Side view of the intersubunit interface at the top layer of the ECD. Residues involved in the interface are shown in stick representation. (C) Side portal in the middle of the ECD between two neighboring subunits. Surface and residues lining the wall are illustrated. (D) TM1-TM2 intersubunit interface. Credit: Science Advances, doi: 10.1126/sciadv.abe5983
Ion permeation pathway and mutagenesis studies
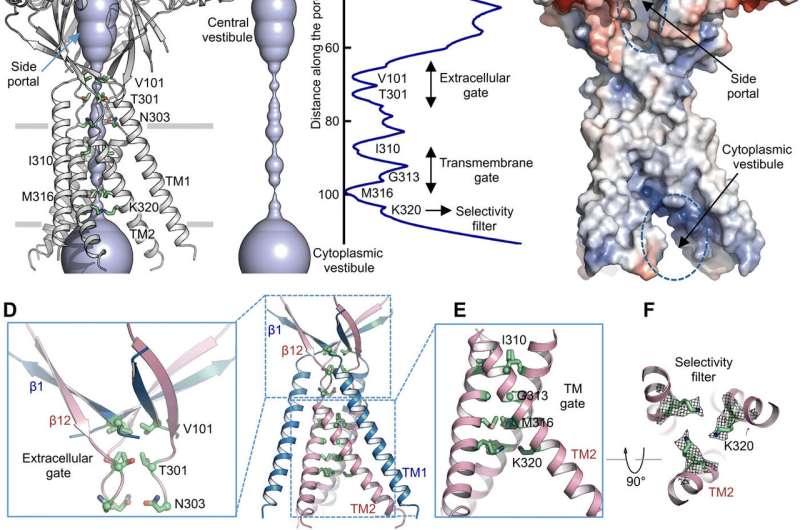
Using pore radius calculations, Deng et al. showed how the central ion conduction pore contained multiple constrictions to prevent ion passage. The structure represented nonconductive conformation due to the high pH buffer conditions (pH 8.0) used to determine the cryo-EM structure. The voluminous and elongated central vestibule could be accessed laterally due to the lack of protein-protein contacts in the extracellular domain. The narrow point of the protein channel did not interfere with ion conduction and could be maintained during the channel gating cycle as ions passed through the three side portals. The slightly positive electrostatic potential of the interior walls of the central vestibule and side entryways also facilitated the attraction of chloride ions. At the extracellular domain-transmembrane domain (ECD-TMD) junction, three strands connected to the outer helices and moved inward to join inner helices to generate an extracellular gate above the bilipid layer. Consecutive constructions at the pore-facing positions formed a hydrophobic gate which could prohibit ion conduction.
To support the structural findings, Deng et al. performed mutagenesis studies on key pore-living residues. When they substituted the conserved basic residues with alanine or acidic residues, the acid-activated chloride currents were abolished. In contrast, arginine substitutions retained channel function to further support the requirement of positive charges in the anion-selective filter. For instance, the I310 domain formed a critical component of the transmembrane gate and another structure known as K320 formed the anion selectivity filter. The work supported structural conservation between the human and pufferfish orthologs to represent a physiologically relevant model for chloride channels.
Ion permeation pathway. (A) Structure of TMEM206EM and the central ion conduction pore, shown in surface representation. Residues generating constrictions are highlighted and labeled. The side portal is indicated. (B) Central ion conduction pore and estimation of the radius (right panel). (C) Surface representation of the channel, colored by surface electrostatic potential (red, −5 kT/e; white, neutral; blue, +5 kT/e). The side portal and cytoplasmic vestibule are indicated. (D) Extracellular gate at the ECD-TMD junction. V101, T301, and N303 are shown in stick representation. (E) Transmembrane gate, constituted by I310, G313, and M316. (F) Putative selectivity filter defined by K320. Also shown are side-chain densities for K320, contoured at 6.5 σ. (G and H) Current densities with an extracellular pH of 7.3 (G) and 4.6 (H) at +100 mV for TMEM206 mutants. The whole-cell membrane currents were recorded by using voltage ramping from −100 to +100 mV for 500 ms at a holding potential of 0 mV. (I) Ratio of current density at pH 4.6 to pH 7.3. Credit: Science Advances, doi: 10.1126/sciadv.abe5983
Structural convergence
The topology, structure and assembly of TMEM206 represented those of the epithelial sodium channel (ENaC)/degenerin superfamily of ion channels, including acid-sensing ion channels (ASICs); despite a lack of distinct amino acid sequence homology between the two channels. The trimeric channels for oppositely charged sodium and chloride ions, nevertheless shared a common core structure flanked by two transmembrane helices. The team noted how the extracellular gate between the central vestibule and transmembrane pore could be expanded to pass ions through after channel activation. While epithelial sodium channels could be activated by releasing inhibitory peptides through proteolysis in the extracellular domain, the ASICs and TMEM206 could only be activated by extracellular protons. The conserved structural features between TMEM and AS1CS also suggested analogous gating conformational changes for the two molecules.
Structural comparison with ASIC and ENaC. (A to C) Subunit structures of TMEM206 (A), ASIC1a [Protein Data Bank (PDB): 6AVE] (B), and ENaC (PDB: 6BQN) (C). Domains are similarly colored. (D) Superposition of TMEM206, colored as in (A), and ASIC1a colored in cyan. (E) Trimeric TMEM206 channel and its central ion conduction pore. The pore is estimated using the program HOLE and depicted as colored dots (pore radius: red < 1.15 Å < green < 2.3 Å < blue). (F) Trimeric ASIC1a channel and the central pore in the closed (middle, PDB: 6AVE) and open (right, PDB: 4NTW) states. Credit: Science Advances, doi: 10.1126/sciadv.abe5983
Outlook
In this way, Zengqin Deng and colleagues used single-particle cryo-EM to determine the structure of integral membrane proteins that are typically unattainable using traditional X-ray crystallography. It is still a significant technical challenge to achieve near-atomic resolution for membrane proteins of small size due to low contrast and signal-to-noise ratios. Deng et al. obtained a 3.5 angstrom resolution structure of a channel by fusing a small crystallization chaperone BRIL to improve stability of the otherwise suboptimal membrane proteins to promote crystal packing. Based on the conserved core structure, the channels were selective for either cations or anions and experienced similar gating conformational changes. The work established a new class of chloride channels to form a new framework for further functional and mechanical investigations in cell and structural biology.
More information: Zengqin Deng et al. Cryo-EM structure of a proton-activated chloride channel TMEM206, Science Advances (2021). DOI: 10.1126/sciadv.abe5983
Veronica Kane Dickson et al. Structure and insights into the function of a Ca2+-activated Cl− channel, Nature (2014). DOI: 10.1038/nature13913
A. Caputo et al. TMEM16A, A Membrane Protein Associated with Calcium-Dependent Chloride Channel Activity, Science (2008). DOI: 10.1126/science.1163518
Journal information: Science Advances , Nature , Science
© 2021 Science X Network
![Structural comparison with ASIC and ENaC. (A to C) Subunit structures of TMEM206 (A), ASIC1a [Protein Data Bank (PDB): 6AVE] (B), and ENaC (PDB: 6BQN) (C). Domains are similarly colored. (D) Superposition of TMEM206, colored as in (A), and ASIC1a colored in cyan. (E) Trimeric TMEM206 channel and its central ion conduction pore. The pore is estimated using the program HOLE and depicted as colored dots (pore radius: red < 1.15 Å < green < 2.3 Å < blue). (F) Trimeric ASIC1a channel and the central pore in the closed (middle, PDB: 6AVE) and open (right, PDB: 4NTW) states. Credit: Science Advances, doi: 10.1126/sciadv.abe5983 Cryo-electron microscopic structure of a proton-activated chloride channel named TMEM206](https://scx1.b-cdn.net/csz/news/800a/2021/4-cryoelectron.jpg)