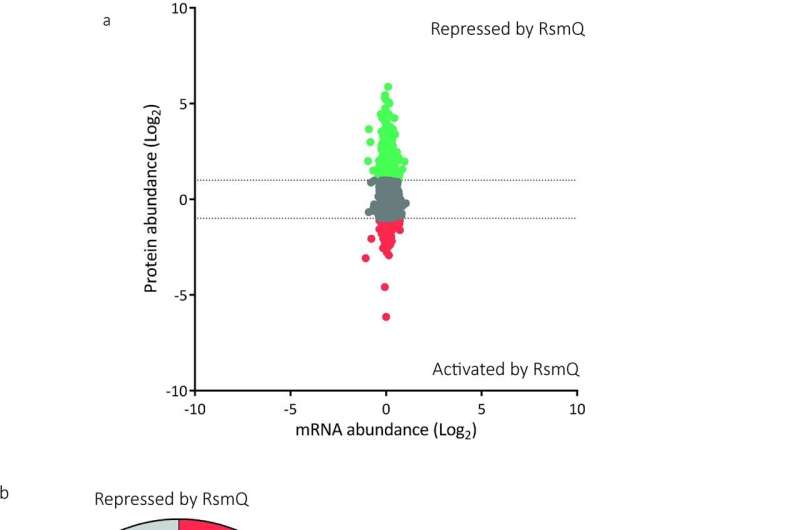
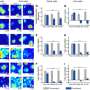
The loss of RsmQ causes widescale proteomic changes. (a) Comparative scatter plot comparing log2-fold mRNA abundance changes from RNAseq (n = 5) to protein abundance changes seen by TMT quantitative proteomics (n = 3). (b) COG categories of proteins that showed a greater than log2-fold change when rsmQ is lost (Repressed by RsmQ = 581, Activated by RsmQ = 203). Data are available in S1 Table and S2 Data. This data is also publicly available in ArrayExpress (E-MTAB-11868) and ProteomeXchange (PXD033640). Credit: PLOS Biology (2023). DOI: 10.1371/journal.pbio.3001988
Bacteria can pass genetic information among themselves to gain an advantage over competitors in their environment.
One of the ways they do this is by transferring conjugative plasmids—mobile, independent pieces of DNA—that often give the bacteria carrying them a beneficial trait such as the ability to survive in the presence of antibiotics or an enhanced ability to colonize plants.
However, sometimes these plasmids can persist for long periods of time in the absence of any obvious advantage to their host. Until now it has not been fully understood why this happens and to what extent plasmids can manipulate gene expression in bacteria.
Research by the Malone group at the John Innes Centre sheds new light on this relationship. This leads to opportunities to better understand plasmid spread and persistence, including insights into how the interaction between pathogens and multi-drug resistance plasmids drives antibiotic resistance in the clinic.
The study, which appears in PLOS Biology, investigates a plasmid gene that encodes a protein called RsmQ.
Experiments using the soil bacterium Pseudomonas fluorescens showed that RsmQ could manipulate how the host bacteria perceive the outside environment. When the plasmid had RsmQ present, the cell responded differently, surviving better with different food sources, and moving less.
The results show that plasmids can regulate and manipulate bacterial behavior to a greater extent than had previously been observed.
"The most exciting thing about this study is seeing a plasmid being able to completely rewire the host cell," explained first author Dr. Catriona Thompson.
"This means that plasmids can move around in a bacterial population and change how their hosts behave, both in the lab and in the soil. This could impact our understanding of how plasmids persist and change bacterial behavior in the environment."
The study reports that RsmQ is the first case of a plasmid-encoded global regulator, a protein able to turn other proteins on and off, changing the behavior of the cell.
"Having these global regulators suggests a much more symbiotic relationship between plasmids and their hosts," explains Dr. Thompson. "This challenges the way we usually think about plasmids, which is that plasmids only persist when they are beneficial to the bacteria."
RsmQ is found on many large conjugative plasmids, including other environmental and clinically relevant plasmids.
If we can understand how this protein can impact plasmid behavior, this has implications for tackling antibiotic resistance in the clinic as well as both crop protection and production.
More information: Catriona M. A. Thompson et al, Plasmids manipulate bacterial behaviour through translational regulatory crosstalk, PLOS Biology (2023). DOI: 10.1371/journal.pbio.3001988
Journal information: PLoS Biology
Provided by John Innes Centre