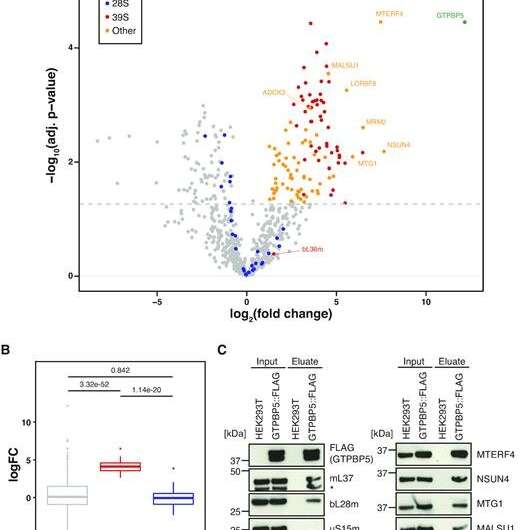
GTPBP5 interacts with the mt-LSU proteins and the MTERF4:NSUN4 in the mitoribosome assembly complex. (A) Quantitative mass spectrometry analysis of proteins interacting with GTPBP5::FLAG. Following FLAG-IP, eluates from HEK293T expressing GTPBP5::FLAG and control HEK293T without FLAG protein expression (WT) were subjected to label-free quantitative mass spectrometry (LFQ) (n = 3). Proteins with a positive log2(fold change) (logFC > 3) and –log10(P-value) of >1.3 (adjusted P-value < 0.05) are identified. Identified proteins are as indicated in the key. A fully annotated version of this dataset is presented as Supplementary Figure S2A and Table S5. (B) Boxplot comparison of the logFC of core proteins of the 39S (mt-LSU) or 28S (mt-SSU) relative to global proteins from dataset in A. Stated P-values indicate pair-wise significance of difference in logFC between global proteins, mt-LSU proteins or mt-SSU proteins as determined via Welch's unequal variances t-test. (C) Immunoblotting of GTPBP5::FLAG-IP. Input mitochondrial lysates and eluate of FLAG-IP from HEK293T expressing GTPBP5::FLAG and control HEK293T without FLAG protein expression (WT) were resolved via SDS-PAGE, western blotting was performed and subsequent membranes were probed with antibodies against FLAG and for proteins of either the mt-LSU (bL28m, mL37) or mt-SSU (uS15m, uS17m) (left panel), or for known mt-LSU assembly factors (right panel). The extra band in mL37 panel (*) represents NSUN4 signal from previous probing. Credit: Karolinska Institutet
Researchers at Karolinska Institutet characterize a GTPase involved in ribosome biogenesis in mitochondria. The study, published in Nucleic Acid Research, reveals new details of a complex process that enables mitochondria to produce energy.
Human cells contain two types of ribosomes, the cell's so-called protein factories. One exists in the cytosol, the other in the mitochondria. In mitochondria, mitoribosomes synthetize the small subset of proteins responsible for oxidative phosphorylation and energy production. Their dysregulation contributes to complex pathologies, including neurodegenerative diseases, such as Alzheimer's and Parkinson's disease.
Building mitoribosomes requires coordinated action of multiple protein factors
The synthesis of ribosomes is an extremely complex, multistep process, which includes both ribosomal RNA (rRNA) folding and the ordered association of dozens of accessory proteins to the growing ribosome. This complexity explains why many of the details of the process are incompletely understood.
In bacteria, who are hypothesized to share evolutionary history with mitochondria, several GTP-binding proteins have been shown to be involved in ribosome assembly.
Two human homologues of the bacterial assembly factor ObgE have been found to reside in human mitochondria, GTPBP10 and GTPBP5. The function of GTPBP10 has previously been investigated by the authors of this study, revealing it to be involved in the assembly of the mitoribosomal large subunit. In the current study, the researchers investigate GTPBP5. Using proteomic and biochemical analyses, they show that GTPBP5 interacts with several previously characterized assembly factors during latest stages of mitoribosome production. Loss of the factor results in impaired mitochondrial translation, making its function of fundamental importance to cellular metabolism.
"Understanding the molecular basis underlying complex pathways such as mitochondrial biogenesis is a great challenge. GTPBP5 is only one of many factors implicated in this process and we have only just begun to scratch the surface." says Miriam Cipullo, author and Ph.D. student in the group.
"Our study provides insight into the molecular mechanisms of mitoribosome biogenesis and places GTPBP5 amongst candidate disease genes in patients with OxPhos and other metabolic deficiencies," says Joanna Rorbach, Assistant Professor at the Department of Medical Biochemistry and Biophysics at Karolinska Institutet.
More information: Miriam Cipullo et al. Human GTPBP5 is involved in the late stage of mitoribosome large subunit assembly, Nucleic Acids Research (2020). DOI: 10.1093/nar/gkaa1131
Journal information: Nucleic Acids Research , Nucleic Acid Research
Provided by Karolinska Institutet