Credit: CC0 Public Domain
The red mold Neurospora crassa belongs to the group of sac fungi and serves researchers as a common model organism that allows a comparatively simple analysis of genetic information and the functions associated with it. The fungus has been the subject of scientific research for decades and its genetic information contained in around 10,000 genes has been completely decoded since the beginning of the 21st century. However, even this intensively studied organism contains components about which little is yet known. These include the so-called eisosomes, certain protein structures that are found in the cell membranes of mycelial fungi, yeast and algae and whose functions are still largely unexplored.
Using this mold as an example, a research team from the Botanical Institute at Kiel University succeeded in identifying central building blocks of the eisosome structure and various proteins associated with them. The work provides initial clues to the possible involvement of the eisosomes in various cellular processes. The results were published recently in the journal Frontiers in Microbiology by researchers from the Department of Botanical Genetics and Molecular Biology headed by Professor Frank Kempken.
The composition of eisosomes
Living cells are surrounded by a barrier, the so-called plasma or cell membrane. This complex membrane system performs various central functions, including the spatial and temporal separation of biochemical processes inside and outside cells. In this membrane system, the eisosomes represent a component that has been little studied to date and is composed of various previously unknown proteins. In order to identify their building blocks, the Kiel research team used a method that marks the proteins associated with the eisosomes. The scientists were then able to identify them using mass spectrometry.
Qin Yang, a doctoral student in Kempken's department, succeeded in describing a total of six different components of the eisosomes and five other proteins in N. crassa that are spatially and temporally associated with the eisosomes. These results also allowed the scientists to show differences to the eisosomes of other organisms. "Certain proteins known from yeasts are not found in the fungus. On the other hand, N. crassa contains hitherto unknown and specific proteins, for example the so-called glucosamine-fructose-6-phosphate aminotransferase," says Kempken, member of Kiel University's priority research area Kiel Life Science (KLS).
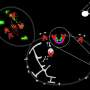
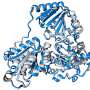
Structural model of the eisosomes
After having determined basic components of the eisosomes, the Kiel researchers used a protein modeling approach and bioinformatic analyses to create an eisosomal structural model for N. crassa. This theoretical spatial depiction of the structure of the eisosomes should in future make it possible to find out more about their functions. "We assume that eisosomes are involved in different cellular mechanisms. It is possible that they contribute to the formation of the cell wall or certain signaling and transport processes in the fungal cells," says Kempken.
The new findings of the Kiel research team suggest that, due to their potential involvement in diverse cellular processes, the eisosomes serve as organizational centers in the plasma membrane. Moreover, they do not appear to form a self-contained structure, but rather consist of a complex interaction of protein groups and independent protein components. Overall, the researchers hope that their modeling of the eisosome proteins will provide inspiration for further research into these membrane components. In the future, it may be possible to draw more attention to this research area and ultimately to elucidate the functional role of the eisosomes in fungi and other organisms.
More information: Qin Yang et al. The Composition and the Structure of MCC/Eisosomes in Neurospora crassa, Frontiers in Microbiology (2020). DOI: 10.3389/fmicb.2020.02115
Provided by Kiel University