Islands in yeast membrane revealed by extreme microscopy
University of Groningen microbiologists have visualized tiny islands in the cell membrane of baker's yeast. These membrane compartments appear to store transport proteins before use. The scientists observed that these proteins move extremely slowly in the plasma membrane of the yeast and discovered how they travel through the membrane to reach the islands. They made these observations with state-of-the-art super-resolution optical microscopy. The results were published in Nature Communications on 5 February.
Until some 10 years ago, textbooks on cell biology would show the cellular membrane as a homogenous lipid bilayer with randomly inserted membrane proteins. But this view changed significantly with the discovery of different phases in the membrane of mammalian cells. The lipid composition can differ on 'islands' in the membrane, which affects protein distribution. However, very little was known about similar structures in yeast cells.
"Electron-microscopy studies from back in the 1960s show depressions in the yeast membrane, but they were not studied in detail and were dismissed as artifacts of sample preparation," explains University of Groningen Professor of Biochemistry Bert Poolman. "Then, about 10 years ago, these depressions were rediscovered. Scientists observed banana-shaped proteins attached to the inner side of the membrane that turned out to be responsible for these depressions, which were named eisosomes."
Poolman decided to study these depressions in the yeast membrane for a number of reasons. "We are deeply involved in a project to build a synthetic cell from molecular components. So we need to know a lot about the membrane and how to get our hands on membrane proteins." Furthermore, the eisosomes are the preferential location of a number of transport proteins that the industry partners of Poolman's research group are interested in.
Transporters
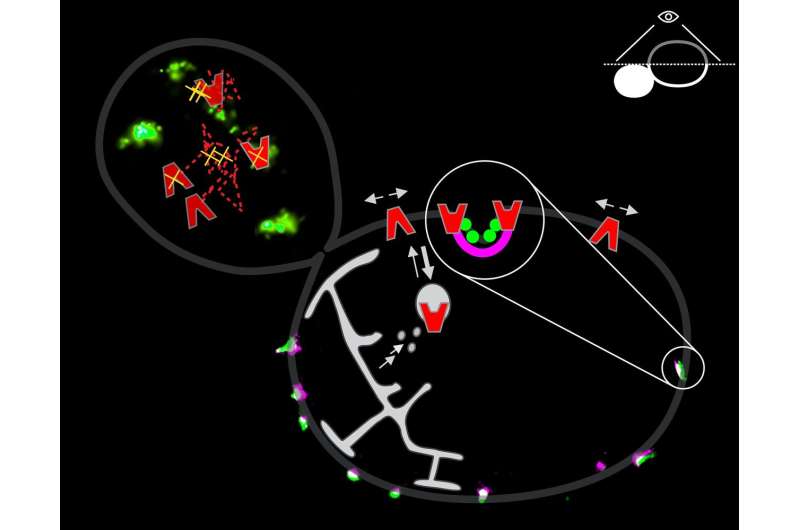
By using different fluorescent markers to label both the transport proteins in the membrane and the banana-shaped proteins on the inner side, the Poolman group determined which proteins are co-localized with the eisosomes. As the depression is only some 50 nanometres deep, and the eisosomes are a maximum of 150 by 100 nanometres in dimension, this required an extremely high resolution. "Fortunately, our lab has a set of dedicated microscopes that can obtain such an extreme resolution, combined with the high sensitivity needed to observe single molecules in living cells." The experience in super-resolution microscopy and expertise in membrane biochemistry allowed the group to provide images with the required resolution.
The studies revealed that some amino acid transporters are preferentially localized in the eisosome. "But only when there is no substrate available," explains Poolman. "If we add the right amino acid, the protein moves away from the eisosome, probably because it takes on a different conformation in the substrate-bound state." His hypothesis is that the eisosomes protect the transport proteins from recycling. "The proteins are synthesized in the cell and then transported to the membrane by exocytosis. However, when they are not in an eisosome, these proteins are quickly absorbed again through endocytosis." So the transporters are transiently 'stored' in the eisosomes. When the appropriate substrates are present outside the cell, they move away to transport the amino acids into the cell until the proteins are no longer needed, after which they are recycled.
Diffusion
Not all proteins are present in the eisosomes. Poolman says, "For example, we noticed that membrane proteins with large intracellular domains cannot enter them." They propose that the banana-shaped proteins on the inner side of the membrane get in the way of the intracellular domains, which hinders their diffusion into the eisosomes.
The Poolman group also assessed the speed of diffusion of the proteins in the yeast plasma membrane. They observed that this was about a thousand times lower than in mammalian cells or in the internal membranes of the yeast cell. "The yeast plasma membrane is more rigid. It can withstand relatively high concentrations of alcohol or acid. This apparently affects protein diffusion."
The results of this study provide a better insight into the functioning of the yeast cell membrane in general, and more specifically the eisosome islands. They also provide new information on the biogenesis and trafficking of membrane transport proteins, which in time may improve the industrial productivity of yeast.
More information: Frans Bianchi et al, Steric exclusion and protein conformation determine the localization of plasma membrane transporters, Nature Communications (2018). DOI: 10.1038/s41467-018-02864-2
Journal information: Nature Communications
Provided by University of Groningen