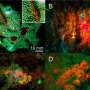
The top and bottom images in column (A) are from the source domain (PVD) and the target domain (PlantDoc dataset). The images in column (B) are apple leaves with rust disease. In column (C), the top and bottom images are respectively potato leaf and tomato leaf with late blight disease. Credit: Plant Phenomics (2023). DOI: 10.34133/plantphenomics.0038
Plant diseases pose a significant threat to nations across the globe, owing to the financial burden they impose and the impact they have on food security. Healthy crops sustain millions of livelihoods, and accurate diagnosis of plant diseases allows for timely interventions to ensure sufficient crop production with minimal yield loss.
Traditional approaches to disease recognition typically follow two paths. The first relies on crop inspection by trained experts, while the second leverages neural networks and image-processing. However, both options have their limitations. While trained experts provide their opinions following an error-prone and time-consuming manual inspection, conventional image-processing methods extract superficial information and need prerequisite training for better predictions.
This implies that it is difficult to make consistent predictions as the complexity of information increases.
In this regard, neural networks–collections of algorithms that detect underlying relationships in data–have delivered promising results in plant disease classification. The caveat is the lack of adequate training data. Good data collection is possible in controlled environments but nontrivial in the real world. In the field, diseases may be rare or not easily observed.
Furthermore, disease samples may have complex backgrounds, varying shapes, and occlusions. Neural network-based disease classifiers are also limited in their ability to apply knowledge to new datasets after training.
Now, a team of researchers has developed a new and relatively simple neural network called the 'Multi-Representation Subdomain Adaptation Network with Uncertainty Regularization for Cross-Species Plant Disease Classification' (MSUN), that accurately classifies plant diseases in natural settings.
(A) to (H) respectively correspond to apple scab leaf, potato leaf late blight, tomato early blight, tomato late blight diseases, corn leaf rust, pepper_bell leaf bacterial spot, potato leaf early blight, and potato leaf early blight. The upper image is the original image of the C-PD dataset, and the lower part is the maps of the network interest areas generated by Grad-CAM for DANN [43] and MSUN, respectively. Credit: Plant Phenomics (2023). DOI: 10.34133/plantphenomics.0038
To do so they applied a transfer learning technique called unsupervised domain adaptation (UDA) to smoothen the process of plant disease identification. The study was led by Associate Professor Xijian Fan from Nanjing Forestry University and was published in Plant Phenomics.
Associate Prof. Fan, who is also the corresponding author of the study, explains, "UDA allowed our model to apply what it had learned during training to a different unannotated dataset. We trained MSUN to classify plant diseases in the controlled environment of a laboratory. It can now use UDA to classify plant diseases in complex field environments."
The team's approach to leveraging UDA in plant disease classification represents a paradigm shift as it overcomes the shortcomings in current UDA-based approaches. First, the images collected in the field are complex–they have several leaves, odd camera shooting angles, and may be blurred. UDA-based classifiers are expected to process this confounding information for accurate disease classification.
Second, these classifiers are unable to make predictions when tackling plants afflicted with varying disease states, infections at different time points, or at multiple sites. Third, the classifiers face a significant challenge when similar disease manifestations may occur. This happens when multiple disease agents infect a single plant species or when a single disease agent infects multiple plant species.
"MSUN is a more capable disease classifier when learning the overall structure of plant-disease features. Also, it captures more details from the information it receives," says Associate Prof. Fan about the new method's advantages. The study found that MSUN was not hampered by the discrepancy that occurs when the same information is collected in a controlled environment versus a field setting.
Importantly, the group validated MSUN's disease classification accuracy using multiple complex plant-disease datasets. When tested using data from the PlantDoc, Plant-Pathology, Corn-Leaf-Diseases, and Tomato-Leaf-Diseases databases, MSUN excelled and outperformed the current crop of classifiers.
The group is optimistic about MSUN's prospects, given its ability to process challenging datasets. They are confident that it can overcome the inherent uncertainty of current disease classifiers, and that it will aids future plant pathology research by providing significant insights into the problems of disease recognition.
More information: Xinlu Wu et al, From Laboratory to Field: Unsupervised Domain Adaptation for Plant Disease Recognition in the Wild, Plant Phenomics (2023). DOI: 10.34133/plantphenomics.0038
Provided by NanJing Agricultural University

![(A) to (H) respectively correspond to apple scab leaf, potato leaf late blight, tomato early blight, tomato late blight diseases, corn leaf rust, pepper_bell leaf bacterial spot, potato leaf early blight, and potato leaf early blight. The upper image is the original image of the C-PD dataset, and the lower part is the maps of the network interest areas generated by Grad-CAM for DANN [43] and MSUN, respectively. Credit: Plant Phenomics (2023). DOI: 10.34133/plantphenomics.0038 New machine learning framework for more accurate plant disease diagnosis](https://scx1.b-cdn.net/csz/news/800a/2023/new-machine-learning-f-1.jpg)