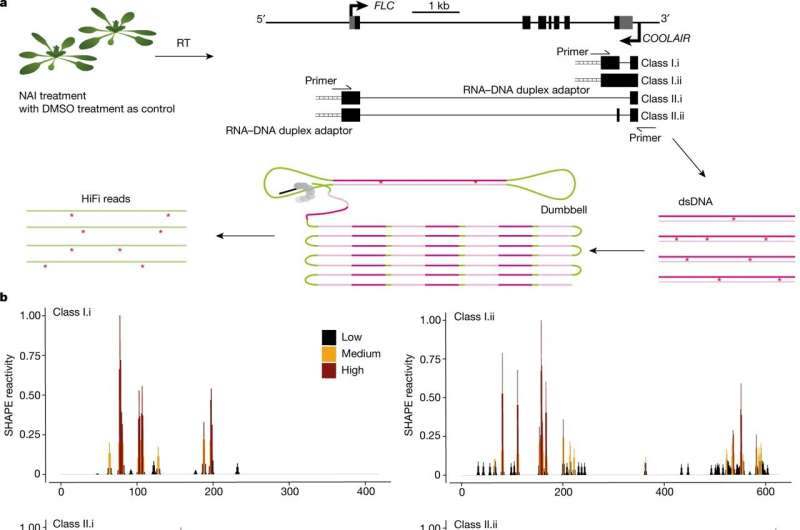
smStructure-seq captures RNA secondary structure information of different transcript isoforms. a, Schematic of the smStructure-seq design for RNA secondary structure probing of each COOLAIR isoform. The Arabidopsis seedlings were treated with NAI ((+)SHAPE) or DMSO ((−)SHAPE). Total RNA was extracted, and the RNA–DNA hybrid adaptors (ladder symbol) were added to the reverse-transcription (RT) reaction using TGIRT-III enzyme. dsDNAs were generated by adding specific primers for all of the COOLAIR isoforms. The dumbbell adaptors were then ligated to the resulting dsDNAs to generate PacBio libraries. The raw subreads were converted to high-accuracy HiFi reads (or circular consensus sequences)14 to generate the mutation rate profiles. b, The normalized SHAPE reactivities derived from the mutation rate profiles were plotted for different class I (under cold-grown conditions) and II (under warm-grown conditions) COOLAIR transcript isoforms. The normalized SHAPE reactivity is calculated from merged n = 2 biological replicates. These reactivity values are color-coded and shown on the y axis. Credit: Nature (2022). DOI: 10.1038/s41586-022-05135-9
The impact of environmental conditions on the dynamic structures of RNAs in living cells has been revealed by innovative technology developed by researchers at the John Innes Centre.
The research, the result of a collaboration between the groups of Professor Dame Caroline Dean FRS and Dr. Yiliang Ding, increases our understanding of what happens at cellular level in response to environmental signals. This raises the possibility that we may use this knowledge to fine-tune crops or develop RNA-based therapies for diseases such as COVID-19 (SARS-COV-2.)
Previous research by these groups showed that two important genetic elements COOLAIR and FLC interplay to regulate plant molecular responses to warm and cold. But it was unclear how the RNA structure of COOLAIR contributes to the regulation of FLC—a genetic brake on flowering in plants.
Researchers in the Ding group developed a new technology which is capable of profiling RNA structure at the resolution of a single molecule in live cells.
Using this technique allowed them to observe RNA structural changes. In warm conditions COOLAIR RNA adopts three predominant structures and these shapes and proportions changed after the plants were exposed to cold temperatures.
They noticed that changes in RNA conformations in one hyper-variable region of COOLAIR changed FLC expression. By introducing mutations into the sequence of this RNA region, the researchers were able to alter the flowering time of the plants.
Dr. Ding says that their "work has shown that RNAs can adopt different conformations, or structures. These diverse conformations dynamically change in response to external conditions. In this study by tuning the RNA structure, we altered the flowering time of the plant."
The understanding of how RNA structure affects RNA function, and the ability to engineer plant genomes at the RNA cellular level, increases the possibility of designing crop types with more desirable agronomic and nutritional traits.
The group says that the technology can also be applied to human cells where RNA structures could serve as a guide for designing RNA-based therapies.
First author Dr. Pan Zhu says that "every RNA is likely to have its own RNA structure landscapes and conformational diversities. Our technology will allow us to explore the pervasive functional importance of RNA structures in the RNAs of interest such as SARS-COV-2."
The group will now look to share their new technology with RNA-based industrial or academic collaborators.
During the process of gene expression DNA is transcribed into RNA which is then used to make proteins. RNA is often referred to as the "skinny molecule" because it is single stranded, but recent work has highlighted its structural diversity and how these structures affect gene regulation and protein synthesis.
In plants FLC acts as a brake on flowering, a key part of a molecular mechanism which ensures the plant only flowers when it has reached a required level of cold exposure. COOLAIR is antisense to FLC, binding to it and blocking it from being transcribed after cold exposure. Knowledge of these mechanisms will be key to understanding the consequences of climate change.
The research appears in Nature.
More information: Caroline Dean et al, In vivo single-molecule analysis reveals COOLAIR RNA structural diversity, Nature (2022). DOI: 10.1038/s41586-022-05135-9. www.nature.com/articles/s41586-022-05135-9
Journal information: Nature
Provided by John Innes Centre