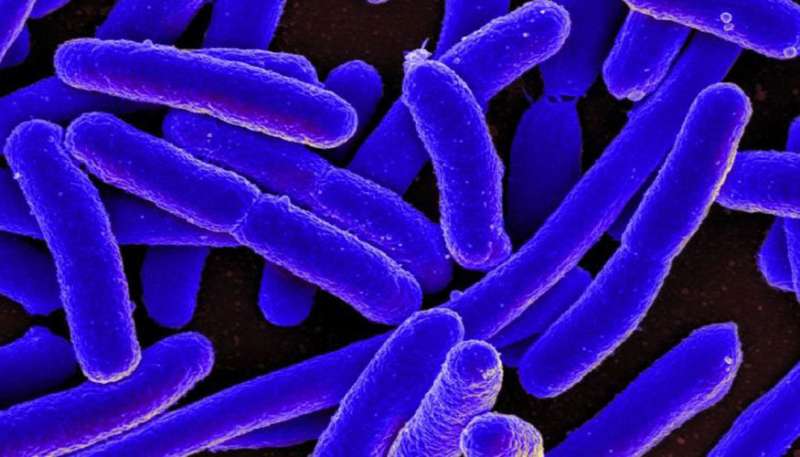
A colorized scanning electron micrograph of Escherichia coli (E. coli). LLNL researchers and their collaborators have developed a new system to control gene expression in laboratory bacteria, including E. coli. Credit: National Institute of Allergy and Infectious Diseases, National Institutes of Health
The microbial production of enzymes, chemicals and fuels could become more efficient and economical with a newly engineered system for controlling genes called "Jungle Express."
Scientists and collaborators from the Department of Energy's (DOE) Joint BioEnergy Institute, Lawrence Livermore National Laboratory (LLNL), Lawrence Berkeley National Lab and San Francisco State University have developed a new system to control gene expression in laboratory bacteria. The research appears in the Sept. 6 edition of Nature Communications.
Bacteria can serve as living factories to produce a wide array of biochemical products. But many bacterial large-scale fermentations have not been efficient before because of the lack of affordable genetic control elements.
"We have created a system that can precisely control the timing and levels of expression of the genes involved in these processes, opening the way for efficient large-scale production at low costs," said LLNL biochemist Michael Thelen, senior author of the paper. "These enhancements to bacterial gene regulation have a potential translation to sustainable bioproduction."
Both in fundamental research and at industrial scale, it is often necessary to maintain strict control over the expression of a gene or set of genes that have been placed into bacterial cells for the biosynthesis of target products. Earlier systems to control gene expression have been in use for many years but have been plagued by one or more key elements. These include "leaky" gene expression, in which biochemical targets are produced too early, frequently leading to undesirable side reactions, toxic products and mutation. In addition, the level of expression is often not high enough to be useful in production. Finally, the reagents used to initiate gene expression are expensive.
The lead author of the article is Thomas Ruegg, a graduate student from Basel University who was working with Thelen. "The key component of this expression system is a DNA binding protein that acts as a gene regulator," Ruegg said. "We used a computational approach to predict a small DNA sequence for optimal binding of the regulator protein."
Ruegg found that the regulator quickly responds to very low concentrations of crystal violet, a readily available and inexpensive dye that is used in a variety of applications. Crystal violet causes the regulator to release its hold on the gene, resulting in active gene expression and target biochemical production. "In a rational approach, we used the optimized DNA binding site to engineer a robust system in which crystal violet acts as the inducer to trigger efficient gene expression," Ruegg said.
"Our findings have the potential to overcome the bottlenecks encountered in earlier systems and opens the way for tightly controlled and efficient gene expression that is not restricted to host organism, substrate or scale," Thelen said.
The name Jungle Express is derived from a rainforest bacterium, the native host of the regulator protein used in this study. Ruegg thinks that "it is a fascinating journey to explore the inexhaustible resource of genetic information found in microorganisms, and to decouple it from its native context for the development of novel useful applications."
More information: Thomas L. Ruegg et al. Jungle Express is a versatile repressor system for tight transcriptional control, Nature Communications (2018). DOI: 10.1038/s41467-018-05857-3
Journal information: Nature Communications
Provided by Lawrence Livermore National Laboratory