May 9, 2024 report
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
AlphaFold 3 upgrade enables the prediction of other types of biomolecular systems
A combined team of medical researchers and AI systems specialists from Google's Deep Mind project and Isomorphic Labs, both in London, has made what the group describes as substantial improvements to AlphaFold 2 that make it possible for the application to predict the structure of a wide variety of biomolecular systems more broadly and accurately. The new iteration is called AlphaFold 3.
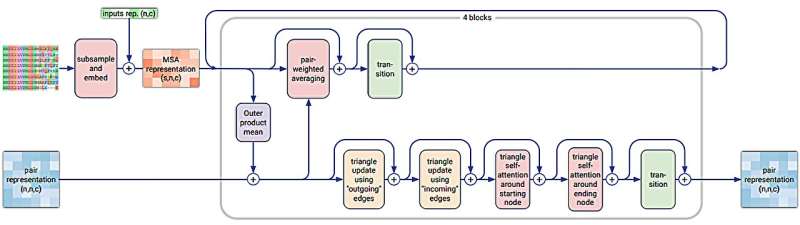
In their study published in the journal Nature, the group used diffusion techniques to make improvements to the underlying architectural model of the application to allow for making more general predictions.
The first version of the deep-learning-based AI system AlphaFold was released just four years ago and was heralded for its ability to make accurate predictions about the structure of proteins using sequences of amino acids. It also has helped researchers better understand how proteins work. AlphaFold 2 built on such capabilities, broadening the complexes that could be predicted.
In this new iteration, the research team has given the application the ability to predict biomolecular systems beyond proteins. It can predict ligands, for example, or RNA or DNA structures. They note that it can even make predictions about the structure of ions, nucleic acids, other proteins and interactions between antibodies and antigens.
These abilities, the researchers note, make it a useful tool for the discovery of new drugs. A drug discovery company (and DeepMind spinoff) is already using the new system to do just that.
In addition to making predictions about other biomolecular structures, the research team claims that AlphaFold 3 is also much more accurate than its previous iterations and its competitors. But they also acknowledge that there is room to grow: AlphaFold 3 has a chirality error rate of 4.4%, for example. It also sometimes hallucinates, which reduces the appearance of ribbons.
They note that work will continue with the AlphaFold system as the team seeks to improve accuracy and add more types of systems to which it can be applied. They also plan to introduce a ranking structure to help users make judgments about results provided by the system.
More information: Josh Abramson et al, Accurate structure prediction of biomolecular interactions with AlphaFold 3, Nature (2024). DOI: 10.1038/s41586-024-07487-w
Journal information: Nature
© 2024 Science X Network