Cells are restless. They move during embryogenesis, tissue repair, regeneration, chemotaxis. Even in disease, tumor metastasis, cells get around. To do this, they have to keep reorganizing their cytoskeleton, removing pieces from one end of a microtubule and adding them to the front, like a railroad with a limited supply of tracks. The EB family of proteins helps regulate this process and can act as a scaffold for other proteins involved in pushing the microtubule chain forward.
Still, how these EB proteins function in space and time has remained a mystery. Now Peng Xia and Xuebiao Yao of the Hefei National Laboratory for Physical Sciences at the Nanoscale and University of Science and Technology of China, and their colleagues, describe in the December 11 issue of Molecular Biology of the Cell (MBoC) how they managed to visualize protein interactions at nanometer spatial resolution in live cells. Yao will also present at the 2014 ASCB/IFCB meeting in Philadelphia on December 8 at 1:30 pm in the ASCB Learning Center. The new method uses photoactivatable complementary fluorescent proteins (PACF) to observe and quantify protein-protein interactions in live cells at the single molecule level.
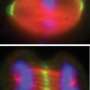
Through clever biochemistry combined with superresolution imaging techniques similar to those that won this year's Nobel Prize in Chemistry, Yao and Xia introduced two EB proteins into cells, one with half of a photoactivatable green fluorescent protein (PAGFP), and one with the other half of PAGFP. These complementary PAGFP pieces will only fluoresce if the EB proteins are in a complex together and photoactivated. They can also be switched off with a different wavelength of light. By activating and then bleaching subsets of EB molecules, the researchers could assemble super-resolution images of protein complexes.
Yao and Xia say their technique has already revealed a surprisingly critical role for a previously uncharacterized EB1 linker region in tracking microtubule plus-ends in live cells. The technique offered precise localization of dynamic microtubule plus-end hub protein EB1 dimers, and their distinct distributions at the leading edge and cell body of migrating cells, the researchers report. And their technique can be applied to the study of other protein complexes in unprecedented detail.
More information: Mol. Biol. Cell. October 29, 2014 mbc.E14-06-1133. DOI: 10.1091/mbc.E14-06-1133
Journal information: Molecular Biology of the Cell
Provided by American Society for Cell Biology