Scientists have cracked a 35-year-old mystery about the workings of the natural motors that are models for development of a futuristic genre of synthetic nanomotors that pump therapeutic DNA, RNA or drugs into individual diseased cells. Credit: Zhengyi Zhao
Scientists have cracked a 35-year-old mystery about the workings of the natural motors that are serving as models for development of a futuristic genre of synthetic nanomotors that pump therapeutic DNA, RNA or drugs into individual diseased cells. Their report revealing the innermost mechanisms of these nanomotors in a bacteria-killing virus—and a new way to move DNA through cells—is being published online today in the journal ACS Nano.
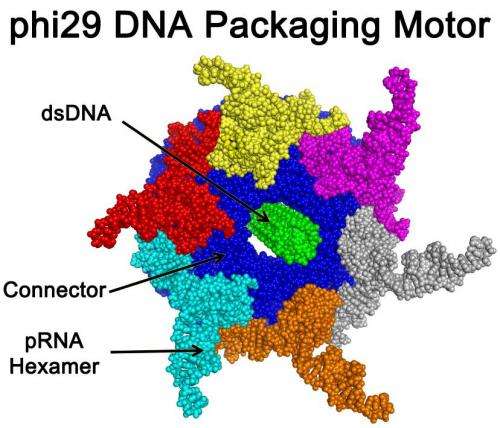
Peixuan Guo and colleagues explain that two motors have been found in nature: A linear motor and a rotating motor. Now they report discovery of a third type, a revolving molecular motor. Guo pointed out that nanomotors will open the door to practical machines and other nanotechnology devices so small that 100,000 would fit across the width of a human hair. One major natural prototype for those development efforts has been the motor that packages DNA into the shell of bacteriophage phi29, a virus that infects and kills bacteria. Guo's own research team wants to embed a synthetic version of that motor into nanomedical devices that are injected into the body, travel to diseased cells and pump in medication. A major barrier in doing so has been uncertainty and controversy about exactly how the phi29 motor moves. Scientists thought that it worked by rotating or spinning in the same motion as the Earth turning on its axis.
In their ACS Nano paper, Guo, with his team Zhengyi Zhao, Emil Khisamutdinov and Chad Schwartz, challenges that idea. Indeed, they discovered that the phi29 motor moves DNA without any rotational motion. The motor moves DNA with a revolving in the same motion as the Earth revolving around the sun. "The revolution without rotation model could resolve a big conundrum troubling the past 35 years of painstaking investigation of the mechanism of these viral DNA packaging motors," the report states.
More information: "Mechanism of One-Way Traffic of Hexameric Phi29 DNA Packaging Motor with Four Electropositive Relaying Layers Facilitating Anti-Parallel Revolution" pubs.acs.org/doi/abs/10.1021/nn4002775
Abstract
The importance of nanomotors in nanotechnology is akin to that of mechanical engines to daily life. The AAA+ superfamily is a class of nanomotors performing various functions. Their hexagonal arrangement facilitates bottom-up assembly for stable structures. Bacteriophage phi29 DNA-translocation motor contains three co-axial rings: a dodecamer channel, a hexameric ATPase ring, and a hexameric pRNA ring. Viral DNA-packaging motor has been believed to be a rotational machine. However, we discovered a revolution mechanism without rotation. By analogy, the earth revolves around the sun while rotating on its own axis. One-way traffic of dsDNA translocation is facilitated by five factors: 1) ATPase changes its conformation to revolve dsDNA within hexameric channel in one direction; 2) the 30° tilt of the channel subunits causes an anti-parallel arrangement between two helices of dsDNA and channel wall to advance one-way translocation; 3) unidirectional flow property of the internal channel loops serves as a ratchet valve to prevent reversal; 4) 5'-3' single-direction movement of one DNA strand along the channel wall ensures single direction; and 5) four electropositive layers interact with one strand of the electronegative dsDNA phosphate backbone, resulting in four relaying transitional pauses during translocation. The discovery of a riding system along one strand provides a motion nano-system for cargo transportation and a tool for studying force generation without coiling, friction, and torque. The revolution of dsDNA among 12 subunits offers a series of recognition sites on DNA backbone to provide additional spatial variables for nucleotides discrimination for sensing applications.
Journal information: ACS Nano
Provided by American Chemical Society