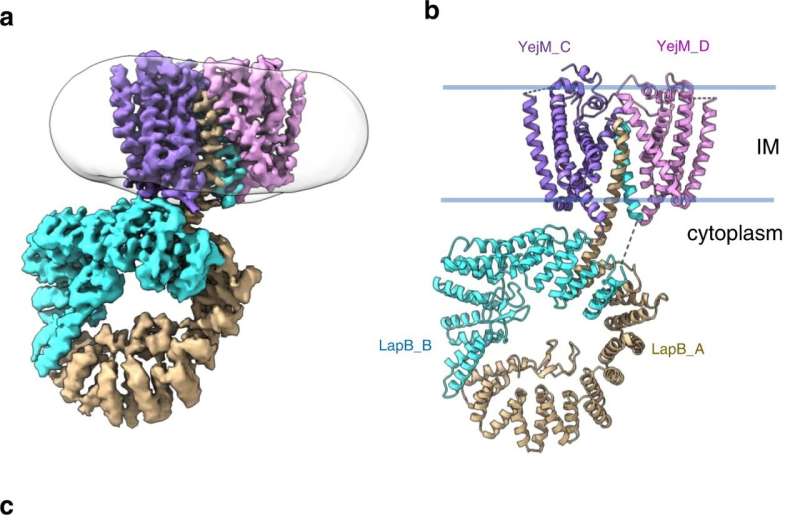
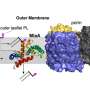
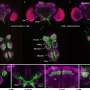
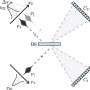
CryoEM structure of the YejM/LapB complex. a Surface view of 3D reconstruction of the YejM/LapB complex, filtered to 3.9 Å resolution. Volumes for proteins are presented with a contour level of 0.25. Molecules of LapB_A, LapB_B, YejM_C, and YejM_D are colored in brown, cyan, purple, and pink, respectively. The volume for the detergent micelle is smoothened with a Gaussian filter and shown as a transparent outline. b Ribbon diagram of an atomic model of the YejM/LapB complex, with the thick blue lines indicating the boundaries of the inner membrane (IM). c Side and top views of the TM domains of the YejM/LapB complex, with a pseudo-two-fold axis shown as a dashed line or a dot. Credit: Nature Communications (2022). DOI: 10.1038/s41467-022-32277-1
Antibiotic-resistant bacteria are making it increasingly difficult to combat many infections. And types of bacteria classified as "Gram-negative"—which refers to the Gram staining method used to differentiate bacteria—are particularly hard to combat as they have an additional protective layer of their cell wall.
Yale researchers have now uncovered a key part of the process that generates this protective layer, a finding that may help reveal vulnerabilities that could be targeted by new antibiotics.
Their findings are published in Nature Communications.
An essential part of this protective outer membrane layer is a molecule called lipopolysaccharide (LPS), previous research has revealed. To survive, bacteria cells need a certain amount of LPS—too much or too little kills the cell. But how bacteria cells determine the right amount remains unclear.
"How do bacteria decide to produce enough LPS to protect them but not so much that it kills them?" said Wei Mi, assistant professor of pharmacology and senior author of the new study.
To answer that question, he and his postdoctoral associate Sheng Shu turned to E. coli.
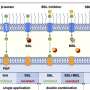
Through a series of experiments, they were able to uncover an important part of the LPS regulatory process in E. coli. It involves a protein that sits in the inner part of the cell membrane and acts as an LPS "sensor." When the cell needs more LPS, like when it's growing and needs to build out its outer membrane, the sensor protein detects this and, with the help of a few other molecules, allows LPS production to continue. LPS is then shipped out to the outer membrane where it's needed.
But when cell growth slows and LPS isn't needed in the same quantities, LPS starts to accumulate in the inner membrane where it then attaches to the sensor protein, knocking off another molecule in the process. That newly freed molecule then stimulates the breakdown of the enzyme that initiates LPS production, which ultimately causes LPS production to slow.
The result is a tight regulation of LPS that is essential for keeping E. coli cells alive. Disrupting this regulation, said Mi, would be an effective way to kill bacteria.
"Understanding this mechanism," he said, "means we can find more ways to develop new antibiotics."
More information: Sheng Shu et al, Regulatory mechanisms of lipopolysaccharide synthesis in Escherichia coli, Nature Communications (2022). DOI: 10.1038/s41467-022-32277-1
Journal information: Nature Communications
Provided by Yale University