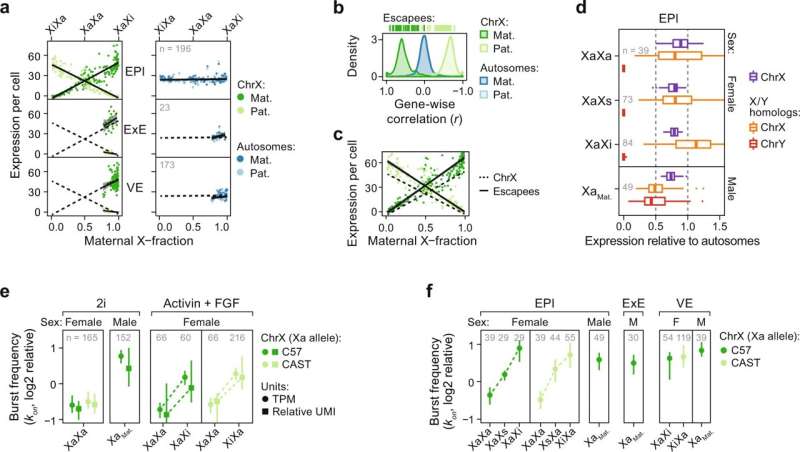
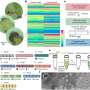
X-upregulation follows a tuning-like mode of regulation. a Scatter plot of expression level per allele (y-axis) and maternal X-fraction per female cell (x-axis) with lines indicating linear model mean ±95% confidence interval. Data are shown for three lineages (EPI, ExE, and VE). Dashed black lines project EPI expression. Shown for autosomes (blue; n = 14,941–17,643 genes) and chrX (green; n = 597–713). b Density plot of gene-wise (n = 712 genes) correlations (Pearson) against maternal X-fractions in EPI cells (n = 196). c Same as a. but shown separately for all X-linked (ChrX) or XCI escapee expression per cell and allele. d X:Autosomal and Y:Autosomal expression ratios per cell shown as box plots grouped by lineage, sex, and XCI state for chrX (n = 129–410 genes) and ancestral X-Y homologs (n = 5–9 genes) separately. X alleles indicated as active (Xa), inactive (Xi), or semi-inactive (Xs). Data are shown as median, first, and third quartiles, and 1.5x IQR. e Transcriptional burst frequency (kon) for active X alleles (Xa per parental strain) grouped by sex, culture condition, and XCI state, shown as median ± 95% confidence interval, inferred by either TPM (dot) or relative UMIs (square). Data are shown relative to median autosomal burst frequency. N genes = 10,836–16,714 and 348–545 for autosomes and chrX, respectively. f Transcriptional burst frequency (kon) of the Xa allele in cells of different lineage, sex, and XCI state in mouse embryogenesis in vivo, shown as median ± 95% confidence interval relative to median autosomal burst frequency, n = 252–316 genes. X alleles indicated as active (Xa), inactive (Xi), or semi-inactive (Xs). Legend as in panel f, female ExE lineage lacked sufficient cells (n ≤ 20) for kinetic inference. a, d–f Number of individual cells per condition shown within plots. Credit: Nature Communications (2022). DOI: 10.1038/s41467-022-29414-1
A new study by researchers at Karolinska Institutet identify major sex differences in the timing of X chromosome upregulation, a finding that is vital for understanding central gene-regulatory processes in early fetal development. The results are published in Nature Communications.
Balanced transcriptional dosage is essential for upholding steady-state gene expression networks in our cells, and how expression dosage is modulated upon gene-copy-number variation is greatly important in biomedicine.
Imbalanced numbers of whole chromosomes result in copy discrepancy of hundreds of genes in a single sweep and is not well tolerated by the cell. Interestingly, while autosomes are present as two active copies the X chromosome stands as an exception in this context, as female cells naturally carry two X's while male cells cope with only one. This challenges the cell as a self-regulating system to rebalance the expression of X correctly in both females (XX) and males (XY), and failure to do so results in embryonic death.
Two major chromosome-wide mechanisms are at play to collectively correct the X dosage in our cells. X inactivation silences one X allele in females, leading to an equalized dosage between the sexes, and X upregulation resolves X-to-autosomal imbalances by increasing expression from the single active X in both sexes. While X-inactivation and upregulation are equally vital to attain correct dosage, the latter has for the most part eluded detailed investigation.
Now, researchers at KI have meticulously characterized these processes in parallel during mouse embryogenesis using techniques capable of quantifying expression of each chromosome copy separately in individual cells.
Elastic process
The study, led by post-doc researcher Antonio Lentini and supervised by principal researcher Björn Reinius at the Department of Medical Biochemistry and Biophysics, identified major sex differences in the timing of X upregulation. Surprisingly, in contrast to the prevailing model of mammalian dosage compensation, in which naïve female cells are initially subject to biallelic X upregulation followed by X inactivation correcting the X dosage, they found that the two processes occur in sync in female cells. Moreover, when blocking the X-inactivation process, X upregulation never initiates.
"We found that X upregulation is an elastic process that actively adjusts expression from one allele in response to the output of the other. It explains how expression levels can remain stable across developmental windows where severe dose imbalance would otherwise be experienced by the female embryo," says lead author Antonio Lentini.
Milestone in X-chromosome regulation
The results are vital for understanding central gene-regulatory processes in the early embryogenesis but also provide keys to the emerging field of regenerative medicine: Reprogramming success of embryonic stem cells tends to be more challenging in females as successful dosage compensation establishment is required.
"Our study represents a milestone in X-chromosome regulation research as it conflicts the conventional dosage compensation model held for decades, adjusting interpretation of hundreds of studies. We provide an improved framework for understanding X-chromosome biology," says Björn Reinius.
More information: Antonio Lentini et al, Elastic dosage compensation by X-chromosome upregulation, Nature Communications (2022). DOI: 10.1038/s41467-022-29414-1
Journal information: Nature Communications
Provided by Karolinska Institutet