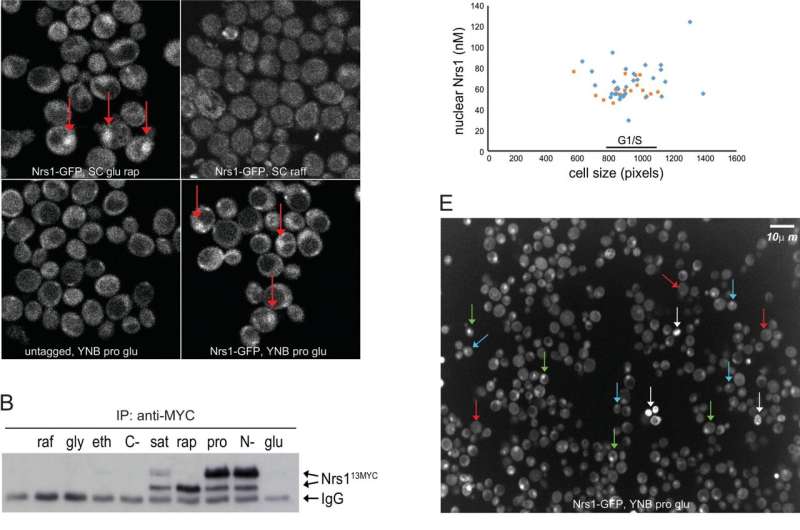
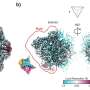
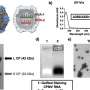
Nitrogen limitation and rapamycin treatment induce cell cycle–regulated Nrs1 expression. (A) sN&B images of untagged and NRS1-GFP strains. Cells were grown and imaged in SC + 2% glucose with or without 100 nM rapamycin, SC + 2% raffinose or nitrogen-limited medium (YNB +0.4% proline + 2% glucose + His, Leu, Met, Ura; labeled YNB+Pro), as indicated. Scale bar is 10 μm. The same intensity scale was used for all conditions. (B) Abundance of Nrs113MYC in various nutrient conditions as determined by immunoblot of anti-MYC immune complexes. raf, 2% raffinose; gly, 2% glycerol; eth, 2% ethanol; C-, no carbon source; sat, saturated culture; rap, 100 nM rapamycin; pro, 0.4% proline; N-, no nitrogen source; glu, 2% glucose. IgG indicates antibody light chain. A raw image of the original immunoblot is provided in S1 Raw Images. (C) Nrs1MYC immunoprecipitates from a rapamycin treatment time course were either mock-treated (M, negative control), treated with lambda phosphatase (P), or lambda phosphatase + phosphatase inhibitors (P+I) prior to detection by anti-MYC immunoblot. In vitro phosphorylated recombinant Sic1 was used as a control to demonstrate activity of the phosphatase. (D) Absolute Nrs1 concentration in single cells grown and imaged in nitrogen-limited medium (YNB+Pro) as a function of cell size, as determined by sN&B. Blue and orange dots represent individual cells from 2 different experiments. The typical size range of cells at the G1/S transition (800 to 1,000 pixels, corresponding to 27 to 38 fL) is indicated. Cell-averaged total Nrs1 concentration (top) and nuclear concentration where Nrs1 nuclear localization was evident (bottom) are shown. Infrequent small cells with high Nrs1 levels had high autofluorescence and no nuclear localization of the signal. (E) Example of high-content confocal image of Nrs1-GFP cells grown to log phase in nitrogen-limited medium. Arrows indicate example cells with strong (green), dim (blue), or no nuclear Nrs1 (red) or inviable cells (white). The histogram summarizes Nrs1 signals in small unbudded (708 cells), large unbudded and small-to-medium budded (599 cells) and medium-to-large budded cells (186 cells) from 18 confocal images. Inviable cells with high autofluorescence, out-of-focus cells for which the budding pattern could not be ascertained and regions in which illumination was not homogeneous were not scored. All numerical values underlying panels D and E may be found in S1 Data. NRS1, Nitrogen-Responsive Start regulator 1; sN&B, scanning Number and Brightness; YNB+Pro, YNB + 0.4% proline + 2% glucose. Credit: PLOS Biology (2022). DOI: 10.1371/journal.pbio.3001548
Researchers from the University of Eastern Finland and the University of Montreal (Canada) have discovered that a micro-protein, which they named Nrs1, supports cell division and proliferation when nutrients are scarce. This result, published in PLOS Biology yesterday, sheds a new light on how evolution subtly reshapes the genomes of unicellular microorganisms, providing them with plasticity to adjust their growth and proliferation to ever-changing environments.
At the turn of the millennium, scientists became able to experimentally determine the DNA sequence of entire genomes. They next used this information to predict genome products: the proteins.
"At this time, very short DNA sections coding for very small proteins were overlooked. Why spend resources studying these tiny, shy dudes when there's already so much to do with the big, tough guys? This strategy paid off to identify central, evolutionary conserved cellular mechanisms; but the adaptation potential, though, hides in less conserved, often short DNA sequences," says University Researcher Sylvain Tollis who carried out the study in Montreal and at the Institute of Biomedicine, University of Eastern Finland.
Furthermore, micro-proteins are increasingly associated with disease in humans: for instance, humanin, which is only 24 amino acids long, is involved in neuronal cell death and survival1, while the cancer-associated microprotein CASIMO1 promotes cell proliferation and motility in breast cancer cell lines through the actin cytoskeleton2. These results urge the community to scrutinize smaller proteins, or micro-proteins, and other genome sequences previously left aside.
In the newly published study, the authors used the bakers' yeast Saccharomyces cerevisiae to seek for molecular routes by which the information on nutrient availability could be communicated to the key molecules, called transcription factors, that orchestrate the commitment to division, referred to as the Start point. Indeed, cell growth and division are strongly affected by the availability of nutrients. For this purpose, they deleted from the yeast genome the main activators of cell division, and overexpressed one by one the remainder of yeast proteins, including many small ones. A unique micro-protein emerged from this screen as capable of rescuing cell proliferation despite the absence of key cell division activators. Further biochemical analyses and ground-breaking quantitative microscopy investigations revealed that cells express this protein only under poor nitrogen conditions, and when it is time to divide. The authors have therefore renamed it as Nrs1 for Nitrogen-Responsive Start regulator. Nrs1 binds to and activates the main transcription factors that trigger the decision to divide, providing an alternative, nutrient-regulated mechanism for Start activation.
Sequence analysis across yeast species indicated that Nrs1 is a recently-evolved microprotein, illustrating how microproteins can rapidly emerge to rewire fundamental cellular processes.
"Indeed, it seems reasonable to assume that short DNA sequences would require less evolution-selected mutations than long sequences to be functionally optimized. This work raises the hypothesis that micro-proteins would make a versatile tool for evolution to quickly rewire key cellular pathways and provide plasticity to adapt to changing environment," Tollis concludes.
More information: Sylvain Tollis et al, The microprotein Nrs1 rewires the G1/S transcriptional machinery during nitrogen limitation in budding yeast, PLOS Biology (2022). DOI: 10.1371/journal.pbio.3001548
Journal information: PLoS Biology
Provided by University of Eastern Finland