This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Protein assembly research shows molecular roll of the dice delivers winning combinations
Australian researchers have shed light on the shape-shifting capabilities of protein assemblies, with results that could revolutionize fields from biomanufacturing to vaccine development.
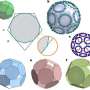
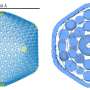
Led by the University of Sydney's Dr. Taylor Szyszka and Dr. Yu Heng Lau, of the ARC Center of Excellence in Synthetic Biology, research published today in the Proceedings of the National Academy of Sciences delves into the intricate world of encapsulins. These icosahedral protein cages play a crucial role in making nano reactors, with their pores acting like doors to tiny nano factories.
"By manipulating the pores and changing their size and charge, we can have better control over substrates entering the cages and being turned into products by the enzymes packaged inside," says Dr. Szyzska.
However, the team's exploration took an unexpected turn when they began making slight mutations to observe the effects.
This roll of the molecular dice led to the creation of structures resembling tetrahedra, a pyramid shape vastly different from the usual spherical encapsulins. In terms of assembly shape, it's akin to changing a 20-sided dice to a four-sided dice. These tetrahedral assemblies, comprising a mere 36 protein subunits compared to the usual 180, open up a realm of possibilities previously unexplored in protein engineering
"What we found is that minimal mutations are required to drastically change the shape of the final assembly," says Dr. Szyszka. "We expected it to be a much more complicated process."
The study's findings not only challenge existing understandings of protein assembly but also offer insights into evolutionary biology. Encapsulins, while not viruses themselves, share evolutionary ties with viral structures. The researchers speculate that the flexibility observed in encapsulins could be attributed to their smaller cargo size compared to viruses, potentially influencing their ability to adopt diverse shapes.
"This discovery suggests that there are other shapes that we can make, with broad implications for bioengineering and biomanufacturing," Dr. Szyszka says.
With further research, the team hope to unlock the full potential of these shape-shifting proteins, paving the way for innovations that could reshape industries and improve human health.
More information: Taylor N. Szyszka et al, Point mutation in a virus-like capsid drives symmetry reduction to form tetrahedral cages, Proceedings of the National Academy of Sciences (2024). DOI: 10.1073/pnas.2321260121
Journal information: Proceedings of the National Academy of Sciences
Provided by ARC Centre of Excellence in Synthetic Biology (CoESB)