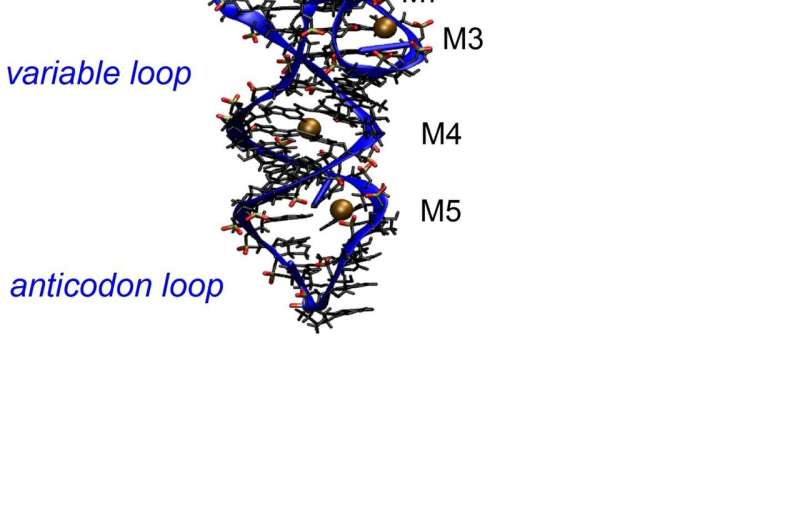
The folded structure consists of a sequence of loop and stem regions. The phosphate-sugar backbone is shown as a blue ribbon to which the nucleobases, shown as molecular structures, are attached. The anticodon loop serves for reading the information which is provided by a messenger RNA and used for synthesizing proteins at the acceptor stem. Contact ion pairs are formed preferentially at the sites M1 to M8. Credit: MBI
In cells, transfer RNA (tRNA) translates genetic information from the encoding messenger RNA (mRNA) for protein synthesis. New results from ultrafast spectroscopy and in-depth theoretical calculations demonstrate that the complex folded structure of tRNA is stabilized by magnesium ions in direct contact with phosphate groups at the RNA surface.
RNA structures consist of long sequences of nucleotides which are composed of a nucleobase, e.g., adenine, uracil, cytosine or guanine, a negatively charged phosphate group, and a sugar unit. The phosphate groups together with the sugars form the backbone of the macromolecule which exists as a folded structure in the cellular environment, the so-called tertiary structure. The tertiary structure of tRNA from yeast has been determined by X-ray diffraction and is shown in Figure 1. For maintaining this structure, a basic prerequisite for its cellular function, the repulsive electric force between the negatively charged phosphate groups needs to be compensated by positively charged ions and by water molecules of the environment. How this works at the molecular level has remained unclear so far, there are conflicting pictures of ion and water arrangements and interactions in the scientific literature.
Scientists from the Max-Born-Institute in Berlin have now identified contact pairs of positively charged magnesium ions and negatively charged phosphate groups as a decisive structural element for minimizing the electrostatic energy of tRNA and, thus, stabilizing its tertiary structure. Their study which has been published in The Journal of Physical Chemistry B, combines spectroscopic experiments and detailed theoretical calculations of molecular interactions and dynamics.
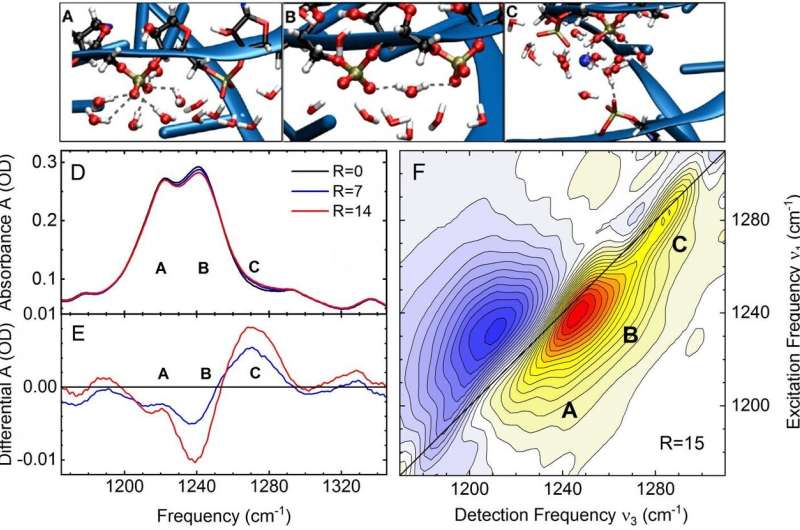
(A-C) Spatial arrangements of phosphate groups (PO4)- (phosphorus: golden, oxygen: red), water molecules H2O (oxygen: red, hydrogen: white) and magnesium ions (blue) as derived from theoretical simulations. The remaining backbone is shown as a blue ribbon. In (A) the phosphate group is surrounded by six water molecules, in (B) by an ordered water structure. In (C) a magnesium ion forms a contact pair with an oxygen of the phosphate group. (D) Infrared absorption spectrum of the asymmetric phosphate stretching vibration of the phosphate group of tRNA for different magnesium concentrations. The quantity R is the ratio of magnesium to tRNA concentration in the sample. The molecular geometries shown in (A-C) result in three different infrared absorption bands A, B and C. With increasing magnesium concentration, the relative strength of component C (contact ion pairs) increases. (E) Differential infrared spectra derived from the data in panel (D). The absorption of contact ion pairs appears as a positive band. (F) Two-dimensional infrared spectrum of tRNA for R=15. The components A, B and C of infrared absorption lead to separate signal contributions (yellow-red contours). The shape of the different bands encodes ultrafast vibrational dynamics of tRNA in its ion/water environment. Credit: MBI
Molecular vibrations of the phosphate groups serve as noninvasive probes of the coupling between tRNA and its aqueous environment. The frequency and infrared absorption strength of such vibrations directly reflects the interactions with ions and water molecules. Vibrational spectroscopy of tRNA samples of different magnesium content together with two-dimensional infrared spectroscopy in the femtosecond time domain allow for discerning specific local geometries in which phosphate groups couple to ions and the water shell (Figure 2). The presence of a magnesium ion in the immediate neighborhood of a phosphate group shifts the asymmetric phosphate stretching vibration to a higher frequency and generates a characteristic infrared absorption band used for detection of the molecular species.
Experiments at different concentrations of magnesium ions show that a single tRNA structure forms up to six contact ion pairs, preferentially at locations where the distance between neighboring phosphate groups is small and the corresponding negative charge density high. The contact ion pairs make the decisive contribution to lowering the electrostatic energy and, consequently, stabilizing the tertiary tRNA structure. This picture is confirmed in a quantitative way by an in-depth theoretical analysis. The ion pairs impose an electrical force on water molecules nearby and orient them in space, again reducing the electrostatic energy. In contrast, mobile ions in the first five to six water layers around tRNA make a smaller contribution to stabilizing tRNA structure.
The new results give detailed quantitative insight in the electric properties of a key biomolecule. They underscore the high relevance of molecular probes for elucidating the relevant molecular interactions and the need for theoretical descriptions at the molecular level.
More information: Jakob Schauss et al, Magnesium Contact Ions Stabilize the Tertiary Structure of Transfer RNA: Electrostatics Mapped by Two-Dimensional Infrared Spectra and Theoretical Simulations, The Journal of Physical Chemistry B (2020). DOI: 10.1021/acs.jpcb.0c08966
Journal information: Journal of Physical Chemistry B
Provided by Forschungsverbund Berlin e.V. (FVB)