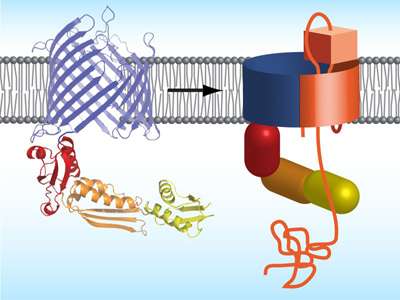
Proposed mechanism how bacteria integrate autotransporter into their outer membrane. Left: protein structure of TamA, right: TamA with autotransporters (orange).
The bacterial outer envelope is densely packed with proteins that form small pores and facilitate the passage of nutrients, toxins and signaling molecules. Professors Timm Maier and Sebastian Hiller from the Biozentrum of the University of Basel now demonstrate how these transporter proteins are integrated into the outer membrane. Using x-ray structural analysis they reveal the structure-function relationship of the protein TamA, which plays an important role in the assembly of transport proteins in the bacterial outer membrane. Their findings have been published recently in the renowned scientific journal Nature Structural and Molecular Biology.
Shuttling proteins from inside the cell to the outside environment is a complex task for Gram-negative bacteria, which are not only surrounded by an inner membrane, but also by an outer membrane barrier for protection against adverse environmental conditions. The bacteria however, can overcome this additional barrier by inserting special transport proteins into the protective outer membrane. In a joint project, Maier and Hiller, both Professors of Structural Biology at the Biozentrum of the University of Basel, provide mechanistic insights into this key process.
The structure of the assembly protein TamA explains its function
An important option for channeling protein domains across the outer membrane are so-called autotransporters. These membrane proteins form a barrel-like structure with a central pore, but they cannot autonomously transport their "passenger domain" across the outer membrane. Specific assembly proteins are required for the folding and integration of autotransporters into the outer membrane. Employing x-ray crystallography, the authors of the study decoded the atomic structure of the autotransporter assembly protein TamA of the intestinal bacterium Escherichia Coli.
"The protein TamA", explains Fabian Gruss, first author and recipient of a Werner-Siemens PhD fellowship, "also forms a barrel with a pore. The pore is closed to the outside by a lid but a particular kink in the barrel wall provides a gate for autotransporter substrates." When an unfolded autotransporter is delivered, TamA hooks onto one end of the substrate polypeptide chain and integrates it step by step via the gate into its own barrel structure. The TamA barrel is thus expanded; the pore widens and opens such that passenger substrates traverse to the exterior. The assembly process ends when TamA releases the autotransporter into the surrounding membrane. "The autotransporter insertion mechanism was previously completely enigmatic – for the first time, knowing the structure of TamA, we can now picture how assembly and translocation could function."
Assembly process important for infections
Many pathogens, such as the diarrhea causing Yersinia, Salmonella or the Cholera pathogen, belong to the group of Gram-negative bacteria. With the help of the autotransporter, they release toxins or adhesive proteins to infect their host cells. In their study, Maier and Hiller provide completely new findings about membrane insertion of autotransporters as well as the translocation of their cargo.
More information: Gruss, F. et al. The structural basis of autotransporter translocation by TamA, Nature Structural and Molecular Biology, 23 September 2013. www.nature.com/nsmb/journal/va … t/abs/nsmb.2689.html
Journal information: Nature Structural and Molecular Biology
Provided by University of Basel