Graphical abstract. Credit: Patterns (2023). DOI: 10.1016/j.patter.2023.100758
The Human Genome Project (HGP), the world's largest collaborative biological project, was a 13-year effort led by the U.S. government with the goal of generating the first full sequence of the human genome. In 2003, HGP produced a genome sequence that accounted for more than 90% of the human genome and was considered as close to complete as was possible with the technologies of the time. HGP unlocked the door to a vast but unannotated collection of genes.
In the following decades, via experimental studies, researchers painstakingly curated reannotations in the form of biochemical reaction graphs. Though gene set enrichment analysis considers groups within these annotation graphs, it disregards group dependencies.
Researchers from the University of Hawaii at Mānoa John A. Burns School of Medicine (JABSOM) are utilizing data from HGP and making advancements in biochemical reaction network analysis. Their work, published in the May 22, 2023 issue of Patterns, demonstrates their approach and may help predict the effects of rare or indistinct genetic variations and guide precision medicine (treatment that can use a patient's own genes to help fight disease or guide specific therapy).
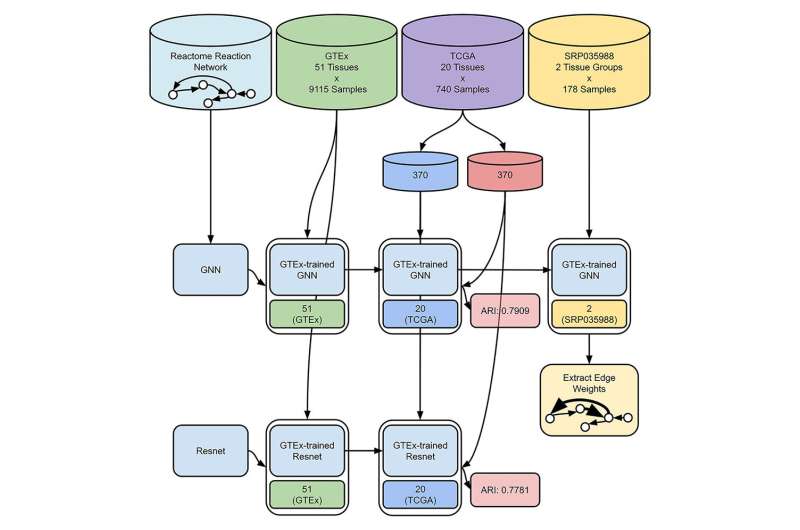
Postdoctoral researcher Joshua G. Burkhart, his supervisor and director Youping Deng, the team in JABSOM's Bioinformatics Core Facility and other co-authors generated a Graph Neural Network based on the Reactome reaction network, a pathway database built using experimental results from prior literature. The Graph Neural Network can find associations, by referencing Reactome connections, that other forms of analysis, such as traditional differential gene expression and hypergeometric enrichment analyses, cannot.
The researchers show how integrating the information or data from this graph with gene expression values from other studies can be used to identify biochemical reactions associated with tissue-specific diseases.
Burkhart and the team demonstrated their model performs comparably to conventional deep learning artificial intelligence. The qualitative benefit of their approach is that additional gene expression datasets may be used to re-tune certain elements in the process which can reveal specific biochemical reactions and subnetworks, traceable to Reactome.
In the future, similar approaches could enable fruitful re-analyses of prior work.
More information: Joshua G. Burkhart et al, Biology-inspired graph neural network encodes reactome and reveals biochemical reactions of disease, Patterns (2023). DOI: 10.1016/j.patter.2023.100758
Journal information: Patterns
Provided by University of Hawaii at Manoa