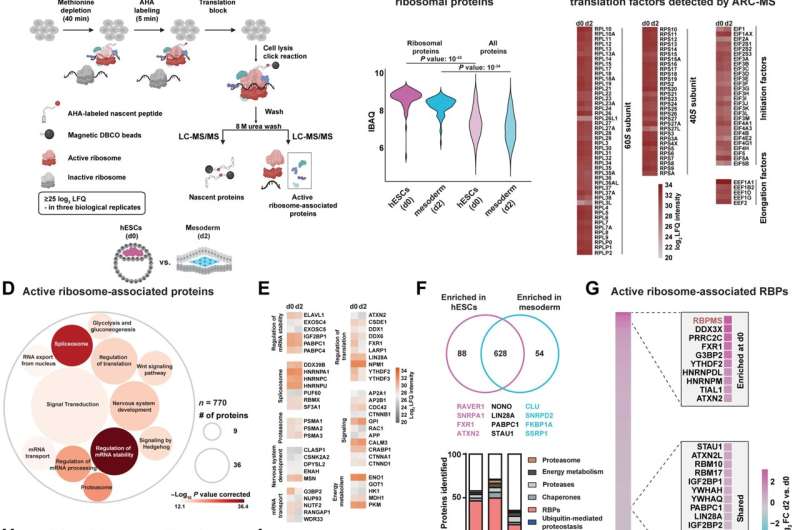
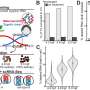
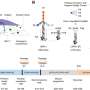
ARC-MS identifies proteins recruited to translationally active ribosomes during mesoderm commitment. (A) Schematic of ARC-MS workflow. ARC-MS was performed in hESCs (d0) and hESC-derived mesoderm progenitors (d2). (B) Violin plots depicting intensity-based absolute quantification (IBAQ) values of ribosomal proteins and all identified proteins from ARC-MS data from hESCs versus mesoderm progenitors. (C) Heatmap showing the enrichment of ribosomal proteins and translation factors (EIFs and EEFs) detected by ARC-MS in hESCs and mesoderm progenitors. (D) GO-based functional enrichment analysis for proteins (excluding ribosomal proteins and translation factors) reliably identified by ARC-MS. (E) Heatmap depicting log2 LFQ of significantly enriched representative proteins recruited on active ribosomes from major GO term categories identified by ARC-MS. (F) Venn diagram summarizing the distribution of proteins on active ribosomes in hESCs and mesoderm progenitors. Identified proteins, categorized based on molecular function, are depicted as a percentage of the total in the bar graphs below. (G) Heatmap displaying the enrichment of RBPs identified by ARC-MS between d0 and d2. (H) Schematic outline of the underlying strategy used for TS-MS. (I) Overlap of proteins enriched at d0 and d2 ARC-MS with proteins detected via TS-MS in hESCs. (J) Distribution of proteins recruited on active ribosomes selectively in hESCs identified by ARC-MS that are overlapping with TS-MS on indicated ribosomal fraction. (K) Confirmation of RBPMS enrichment in ribosomal fractions by polysome profiling followed by immunoblotting. RPL7A, RPS6, and G3BP1 = controls. (L) RBPMS is predominantly a cytosolic protein in hESCs, evaluated by Western blot analysis upon nuclear/cytosolic fractionation, G3BP1, and TUBA cytosolic control, LAMINB1 nuclear control. (M) Residence of RBPMS on ribosomal complexes confirmed by polysome profiling upon treatment with the indicated translation inhibitors. Error bars represent ±SEM; P values are calculated using Student’s t test; biological replicates n = 3. Credit: Science Advances (2023). DOI: 10.1126/sciadv.ade1792
Writing in Science Advances researchers of the University of Cologne describe a key mechanism that controls the decision-making process that allows human embryonic stem cells to make the heart. These discoveries enable better insights into how the human heart forms in an embryo and what can go wrong during heart formation, causing cardiac disease or, in the worst case, embryo termination.
In humans, a specialized mRNA translation circuit predetermines the competence for heart formation at an early stage of embryonic development, a research team at the Center for Molecular Medicine Cologne (CMMC) and the University of Cologne's Cluster of Excellence in Aging Research CECAD led by Junior Professor Dr. Leo Kurian has discovered.
While it is well known that cardiac development is prioritized at the early stages of embryogenesis, the regulatory program that controls the prioritization of the development of the heart remained unclear until now. Kurian and his team investigated how the prioritization of heart development is regulated at the molecular level. They found that the protein RBPMS (RNA-binding protein with multiple splicing) is responsible for the decision to make the heart by programming mRNA translation to approve future cardiac fate choice.
The study is published under the title 'mRNA translational specialization by RBPMS presets the competence for cardiac commitment' in Science Advances.
One out of 100 children born with a cardiac disease
A better understanding of human cardiac development is essential not only to determine the fundamental principles of self-organization of the human heart but also to reveal molecular targets for future therapeutic interventions for congenital and adult-onset cardiac disease.
Since the heart is the first functional organ to form in a developing embryo, any anomaly in early embryonic cell fate decisions needed for the development of the heart leads to catastrophic consequences, often resulting in the termination of pregnancy or lifelong suffering due to congenital heart diseases.
In humans, approximately 30 percent of developing embryos terminate before implantation in the uterus, and about 25 percent fail during the transition from gastrulation (the early phase when the embryo begins to differentiate distinct cell lineages) to organogenesis (the phase that lasts until birth when all tissues and organs form and mature).
Often, the cause of embryo termination is impaired cardiovascular cell fate decisions and morphogenesis, the biological process by which a cell, a tissue or an organism develops its form. The failure to accurately specify cell fate and cell identity in a timely and robust manner results in developmental abnormalities and diseases. For example, 1 out of 100 children are born with congenital cardiac diseases, for the majority of which the causes are unknown.
To discover the regulatory program behind heart development, the Kurian lab used embryonic stem cell-based models that recapitulate human cardiac fate decisions in a dish under chemically defined conditions. The use of human stem cell-derived models allows the team to identify human-specific attributes, which can be drastically different from other animals. The aim of this approach is to work with the most precise models closest to human biology and to minimize animal experiments.
Ribosomes as a regulatory hub to control cellular decision making
The team discovered that the competence for the future cardiac fate is preset in human embryonic stem cells (hESCs) by a specialized mRNA translation circuit controlled by the RNA binding protein RBPMS. RBPMS is recruited to active ribosomes, the molecular machine that produces proteins from mRNA. There, RBPMS controls the production of essential factors needed for the program that triggers the stem cells to develop into heart cells.
Mechanistically, RBPMS has two functions. On the one hand, the protein interacts with components to promote the translation of mRNA to proteins; on the other hand, RBPMS selectively regulates the production of mesoderm signaling components in hESCs by binding to a specific site on the mRNA. The mesoderm is the middle layer of the three germ layers, from which the heart develops early on in embryos.
It is believed that through early contact with cardiogenic signals, the ability of stem cells to develop into future cardiac lineages is predetermined. This study shows that the RBPMS-mediated selective mRNA translation circuit approves the cellular abundance of 'morphogen signaling infrastructure' required for cardiac mesoderm approval in hESCs. Thus, RBPMS sets up the future cardiac competence of hESCs by programming selective mRNA translation.
"In summary, we present a model whereby the state of pluripotency is primed for differentiation into future cell lineages through specialized translation of the regulators of embryonic cell fate. Our work shows that RBPMS selectively programs translation, i.e. the reading of mRNA and the production of proteins or mRNAs. This controls proteins and regulatory mRNAs that themselves code for important developmental regulators and are essential for deciding future cell fate," Dr. Deniz Bartsch, first author of the study, explained.
Based on their findings, the team proposes 'translation specialization': a regulatory mechanism that primes ribosomes to control translation in time and/or space for a set of mRNAs required for future events in response to specific stimuli or fate transitions. This allows efficient division of labor among the approximately ten million ribosomes present in each cell, which are tasked with synthesizing about two million proteins per minute, so the flow of information is streamlined and, as they show, specialized.
This study, therefore, reveals a central role for translational specialization in shaping cell identity during early lineage development and proposes that ribosomes act as a unifying hub for cellular decision-making rather than a mere protein factory.
More information: Deniz Bartsch et al, mRNA translational specialization by RBPMS presets the competence for cardiac commitment in hESCs, Science Advances (2023). DOI: 10.1126/sciadv.ade1792. www.science.org/doi/10.1126/sciadv.ade1792
Journal information: Science Advances
Provided by University of Cologne