Graphical abstract. Credit: Molecular Cell (2022). DOI: 10.1016/j.molcel.2022.06.023
Using an innovative new technique, scientists at Duke-NUS Medical School and their collaborators have identified thousands of previously unknown DNA sequences in the human genome that code for microproteins and peptides potentially critical to human health and disease.
"Much of what we understand about the known 2% of the genome that codes for proteins comes from looking for long strands of protein-coding nucleotide sequences, or long open reading frames," explained computational biologist Dr. Sonia Chothani, a research fellow with Duke-NUS' Cardiovascular and Metabolic Disorders (CVMD) Program and first author of the study. "Recently, however, scientists have discovered small open reading frames (smORFs) that can also be translated from RNA into small peptides, which have roles in DNA repair, muscle formation and genetic regulation."
Scientists have been trying to identify smORFs and the small peptides they code for, since disruption in these smORFs can cause disease. However, currently available approaches are very limited.
"Much of the current datasets do not provide information that is detailed enough to identify smORFs in RNA," added Dr. Chothani. "The majority also comes from analyses of immortalized human cells that are propagated—sometimes for decades—to study cell physiology, function and disease. However, these cell lines aren't always accurate representations of human physiology."
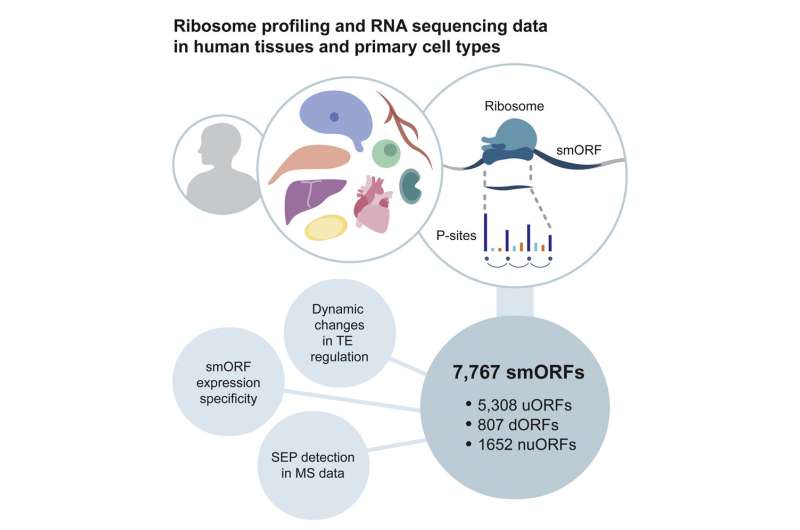
Publishing in Molecular Cell, Chothani and her colleagues in Singapore, Germany, the U.K. and Australia describe a methodology they developed to address these issues. They screened currently available ribosome profiling datasets for short strands of RNA with periodic three-base sections, covering more than 60% of the RNA's length. They then conducted their own RNA sequencing and Ribosome profiling to generate a combined data resource of six types of cells and five types of tissue, such as from the heart and the brain, derived from hundreds of patients.
Analyses of these data identified nearly 8,000 smORFs. Interestingly, they were highly specific to the tissues that they were found in, meaning that these smORFs may perform a function specific to their environment. The team also identified 603 microproteins coded by some of these smORFs.
"The genome is littered with smORFs," said Assistant Professor Owen Rackham, senior author of the study from the CVMD Program. "Our comprehensive and spatially resolved map of human smORFs highlights overlooked functional components of the genome, pinpoints new players in health and disease and provides a resource for the scientific community as a platform to accelerate discoveries."
Professor Patrick Casey, senior vice-dean of research at Duke-NUS, said, "With the healthcare system evolving to not only treat diseases but also prevent them, identifying potential new targets for disease research and drug development could open avenues to new solutions. This research by Dr. Chothani and her team, published as a resource for the scientific community, brings important insights to the field."
More information: Sonia P. Chothani et al, A high-resolution map of human RNA translation, Molecular Cell (2022). DOI: 10.1016/j.molcel.2022.06.023
Journal information: Molecular Cell
Provided by Duke-NUS Medical School