Deeply mining a universe of peptides encoded by long noncoding RNAs
Long noncoding RNAs (lncRNAs), a family of non-coding RNAs (ncRNAs) that are greater than 200 nucleotides in length, were formerly regarded as "junk RNAs" due to the lack of long or conserved open reading frames (ORFs).
Growing amount of evidence has demonstrated that many short or small open reading frames (smORFs) embedded in lncRNA transcripts are able to encode functional polypeptides (smORFs-encoded polypeptides, SEPs). Thousands of additional lncRNA transcripts with smORFs have been discovered, suggesting that SEPs may represent a large albeit neglected portion of non-annotated peptides involved in diverse physiological process. Therefore, large-scale discovery and functional characterization of unknown SEPs might provide new clues for the annotation and functional analysis of noncoding elements in the genome and their effects on biological evolution.
In a study published in Molecular and Cellular Proteomics, Prof. Yang Fuquan's group and Prof. Chen Runsheng's group from the Institute of Biophysics of the Chinese Academy of Sciences implemented a novel strategy for SEP discovery and characterization, which enabled the discovery of 762 novel SEPs from different human and murine cell lines and tissues, representing the largest number of MS-detected SEPs ever to be reported.
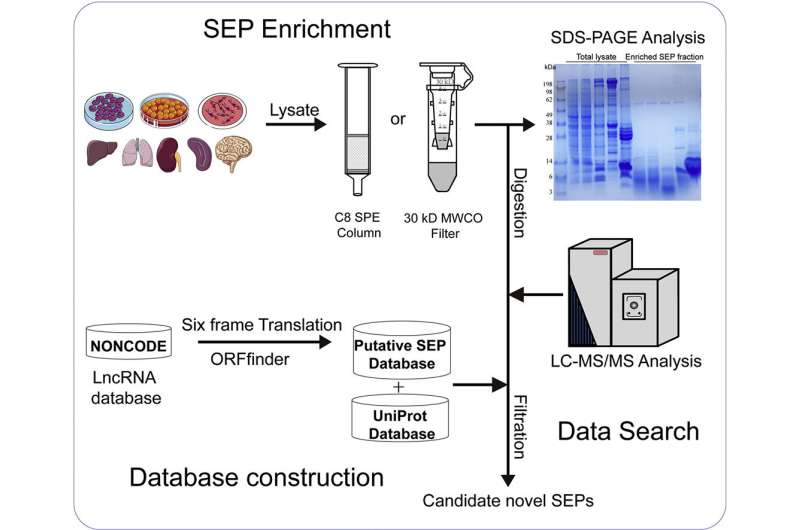
The improved SEP discovery rate can be attributed to an optimized MS-based workflow by combining a de novo construction of a high-quality putative SEP database with two effective and complementary polypeptide enrichment methods.
The researchers take advantage of NONCODE, a repository containing the most complete collection and annotation of lncRNA transcripts from different species, to build a novel database that maximize a collection of SEPs from human and mouse lncRNA transcripts. The combination of 30 kD MWCO membrane filtration and C8 SPE for SEP enrichment also promote the discovery and identification of novel SEPs from different cell cultures and tissues.
The increased number of discovered SEPs in this study provides new opportunities to gain deeper insights into their physical and chemical properties.
For instance, bioinformatic analysis revealed that the physical and chemical properties of these novel SEPs were varied from canonical proteins. More than 60% of the identified SEPs from both human or mouse are initiated by unknown start codons (i.e. non-AUG), which typically have reduced efficiency when compared with AUG codons. Moreover, SEPs are commonly shorter in amino acid length and enriched with more basic residues, which are typical features of known coding genes lacking mass spectrometry evidence.
This study provides new clues for the annotation of noncoding elements in the genome, and serves as a valuable resource for the functional characterization of individual SEPs and the exploration of the possible mechanism of lncRNA translation.
More information: Qing Zhang et al, Deeply Mining a Universe of Peptides Encoded by Long Noncoding RNAs, Molecular & Cellular Proteomics (2021). DOI: 10.1016/j.mcpro.2021.100109
Journal information: Molecular & Cellular Proteomics , Molecular and Cellular Proteomics
Provided by Chinese Academy of Sciences