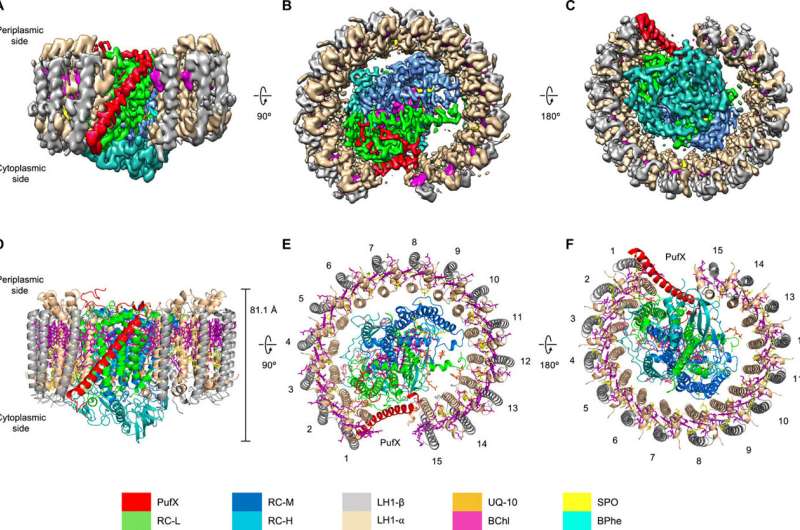
Fig. 1 Cryo-EM structure of the RC-LH1-PufX core complex from Rba. veldkampii. (A to C) Color-coded electron density map in three different views. Color scheme is presented in the legend at the bottom as follows: LH1-α, wheat; LH1-β, gray; PufX, red; RC-L, green; RC-M, marine; RC-H, teal; BChls, purple; BPhes, cyan; carotenoids, yellow; and quinones, orange. (A) Side view of the RC-LH1-PufX complex in the membrane plane. (B) Top view of the RC-LH1-PufX complex from the periplasmic side. (C) Bottom view of the RC-LH1-PufX complex from the cytoplasmic side. (D to F) Architectural model of the RC-LH1-PufX complex in three different views corresponding to (A to C). The height of the RC-LH1-PufX complex perpendicular to the membrane bilayer is shown in (D). The LH1 subunits are numbered in (E) and (F). Credit: Science Advances, 2021, 7, eabf8864
Scientists from the University of Liverpool have uncovered the atomic structure of a special photosynthetic supercomplex to determine how it forms and performs efficient electron transfer.
The study features on the cover of the latest edition of the journal Science Advances.
Purple bacteria are single-cell microorganisms and represent one of the earliest life forms on Earth. These microscopic organisms use sunlight as their source of energy, taking advantage of efficient photosynthetic apparatus located in their photosynthetic membrane. The bacterial photosynthetic apparatus is made of a series of pigment-protein complexes. Through these pigment-protein complexes, solar energy is captured and then converted into chemical energy to power all the functions of the cell.
The central component of bacterial photosynthetic apparatus is a RC–LH1 core supercomplex, which is formed by a light-harvesting 1 ring (LH1) surrounding the reaction center (RC). LH1 captures the photons of sunlight and funnels them to the RC, where the solar energy is converted to chemical energy.
Using high-resolution cryo-electron microscopy (cryo-EM), the research shows a special RC–LH1 structure from a purple photosynthetic bacterium Rhodobacter veldkampii. Unlike a closed LH1 ring found in many purple bacteria, this RC–LH1 core monomer has a "C-shaped" LH1 ring with a large opening. The study further discovered that this opening is formed by an extra protein peptide called PufX. PufX works as a molecular "cross brace" to bridge the RC and LH1 and stabilize the RC–LH1 core structure.
Fig. 2 The PufX structure and interactions within the RC-LH1-PufX complex. (A and B) Schematic model of the RC-LH1-PufX complex represented by cylinders. PufX (red cylinder) exhibits a tilt angle of 43° to the membrane plane (A) and an angle of 68.5° to the orientation of LH1-15αβ heterodimer (B). (C) Color-coded protein surface representation of the RC-LH1-PufX complex. Color scheme is as depicted in Fig. 1. (D) Cartoon representation of the RC-LH1-PufX complex. Interaction sites between PufX and the RC-LH1 assembly are boxed. (E) The interaction network between PufX and RC-L subunit [box 1 in (D)]. Selected PufX residues that interact with RC-L are shown in orange. Selected RC-L residues that interact with PufX are colored green and shown in sticks. All the interacting residues involved in the association between PufX and the RC-L subunit are listed in table S3. (F) The interactions between PufX and LH1-1α and LH1-15α on both sides of the gap in the LH1 ring [boxes 2 and 3 in (D)]. Interacting residues are shown in sticks. (G) The interactions between PufX and LH1-1β [(box 3 in (D)]. Interacting residues are shown in sticks. (H) The interface between PufX and pigments within the LH1-1αβ heterodimer, formed by hydrophobic interactions. Interacting residues are shown in sticks and colored orange if they are interacting with the carotenoid, purple if interacting with BChl, and magenta if interacting with both. PDB ligand ID: BChl, BCL; spheroidene, SPO; and UQ-10, U10. Credit: Science Advances, 2021, 7, eabf8864
The PufX-formed "gate" in this RC–LH1 core supercomplex is the largest among the reported photosynthetic RC–LH1 structures. It provides a big channel for the electron carrier molecules called quinones to diffuse across the LH1 barrier for efficient electron transport. The researchers further used computational approaches to confirm the exchange of quinones through the specific channel.
"Nature has created various intriguing photosynthetic RC–LH1 architectures in ecologically diverse purple bacteria," said Professor Luning Liu at the University of Liverpool, who led the research. "This structural variation represents the distinct mechanisms of energy transfer and electron transport adopted by the photosynthetic apparatus. It allows bacteria to survive in their specific niche. Learning how native photosynthetic complexes are formed and fulfil functions is important for understanding the photosynthetic processes and physiological regulation. We will thus have better solutions to improve photosynthesis and underpin bioenergy production."
More information: Laura Bracun et al, Cryo-EM structure of the photosynthetic RC-LH1-PufX supercomplex at 2.8-Å resolution, Science Advances (2021). DOI: 10.1126/sciadv.abf8864
Journal information: Science Advances
Provided by University of Liverpool
![Fig. 2 The PufX structure and interactions within the RC-LH1-PufX complex. (A and B) Schematic model of the RC-LH1-PufX complex represented by cylinders. PufX (red cylinder) exhibits a tilt angle of 43° to the membrane plane (A) and an angle of 68.5° to the orientation of LH1-15αβ heterodimer (B). (C) Color-coded protein surface representation of the RC-LH1-PufX complex. Color scheme is as depicted in Fig. 1. (D) Cartoon representation of the RC-LH1-PufX complex. Interaction sites between PufX and the RC-LH1 assembly are boxed. (E) The interaction network between PufX and RC-L subunit [box 1 in (D)]. Selected PufX residues that interact with RC-L are shown in orange. Selected RC-L residues that interact with PufX are colored green and shown in sticks. All the interacting residues involved in the association between PufX and the RC-L subunit are listed in table S3. (F) The interactions between PufX and LH1-1α and LH1-15α on both sides of the gap in the LH1 ring [boxes 2 and 3 in (D)]. Interacting residues are shown in sticks. (G) The interactions between PufX and LH1-1β [(box 3 in (D)]. Interacting residues are shown in sticks. (H) The interface between PufX and pigments within the LH1-1αβ heterodimer, formed by hydrophobic interactions. Interacting residues are shown in sticks and colored orange if they are interacting with the carotenoid, purple if interacting with BChl, and magenta if interacting with both. PDB ligand ID: BChl, BCL; spheroidene, SPO; and UQ-10, U10. Credit: Science Advances, 2021, 7, eabf8864 Intriguing structure revealed of a photosynthetic supercomplex in bacteria](https://scx1.b-cdn.net/csz/news/800a/2021/intriguing-structure-r-1.jpg)