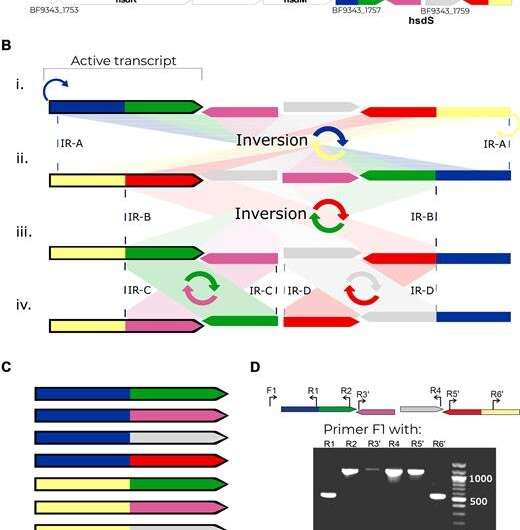
A phase-variable Type-I R-M system in B. fragilis. (A) Gene map of the region. hsdR – restriction enzyme; hsdM- methylase; The colored region of BF9343_1757-60, represents the phase-variable inverting regions of hsdS (specificity genes). (B) Genetic regions showing the specificity genes. i-iv show the different DNA inversions that can occur in this region between inverted repeats shown as dashed lines. Only the outlined gene is transcribed. (C) The eight specificity gene combinations possible from the different DNA inversions. (D) EtBr-stained agarose gel of the amplicons detecting each of the six specificity half genes in the expression locus. Each fragment was amplified using primer F1 paired with each of the six reverse primers.
How does our gut respond and adapt to changing conditions? Where does this fundamental and critical flexibility come from? Technion scientists are unraveling the genius of the gut's microbiome, through microbiota, all the way to genetic inversion.
Assistant Professor Naama Geva-Zatorsky and doctoral student Nadav Ben-Assa of the Rappaport Faculty of Medicine, in collaboration with scientists from Harvard University, have decoded a reversible genetic inversion mechanism that helps a bacterial species of the gut microbiota deal with changes in its habitat. Their findings are published in Nucleic Acids Research, a peer-reviewed scientific journal of Oxford University Press.
The human microbiota refers to the collection of microbes (bacteria, viruses, etc.) that colonize the inner and outer surfaces of the human body. The human intestine contains the most abundant and diverse microbiota population.
Gut microbiota provide a fundamental coping mechanism within the dynamic environment of the gut, in which structural, mechanical, and chemical change occurs incessantly. One mechanism that helps the gut microbiota perform involves rapid, reversible changes in genomes in response to external stimuli.
The article published in Nucleic Acids Research discusses this mechanism in one of the most abundant bacterial species in the human gut, Bacteroides fragilis. This bacterium is capable of inverting a large number of defined regions throughout its genome sequence. The researchers focused on the relationship between this capability and the organism's gene expression.
The research team examined the gene expressions of these changes (recombination) and found extensive alterations in the bacterial genome.
"Among other things, we discovered changes in the sugars surrounding the bacterium," said Prof. Geva-Zatorsky. "These sugars serve as a kind of 'identity card' that helps the bacterium communicate with the environment. With these sugars, they also help our bodies, or more precisely, our immune system, to identify the type of bacterium present, and to respond to it. This is why we assume that changes in the gut alter that 'identity card,' which enables our cells to respond to the bacterium in different ways."
The researchers emphasized that these are reversible genetic inversions, based on recombination of regions in the genome in a major system in the B. fragilis organism. Consequently, this recombination has an extensive effect on the organism's gene expression, including various vital molecules.
Genetic analysis was performed using SMRT (single molecule real-time) sequencing—an innovative technology from Pacific Biosciences (PacBio) developed in the past decade. The technology enables the long-range sequencing and mapping of DNA molecules, as well as the detection of epigenetic DNA modifications. In the system that was researched, genetic recombination affected genetic modifications and consequently the gene expression of B. fragilis in its entirety. The system can also detect hostile elements such as bacteriophages, and this is the subject of a new research study that is now underway in the laboratory.
More information: Nadav Ben-Assa et al, Analysis of a phase-variable restriction modification system of the human gut symbiont Bacteroides fragilis, Nucleic Acids Research (2020). DOI: 10.1093/nar/gkaa824
Journal information: Nucleic Acids Research
Provided by Technion - Israel Institute of Technology