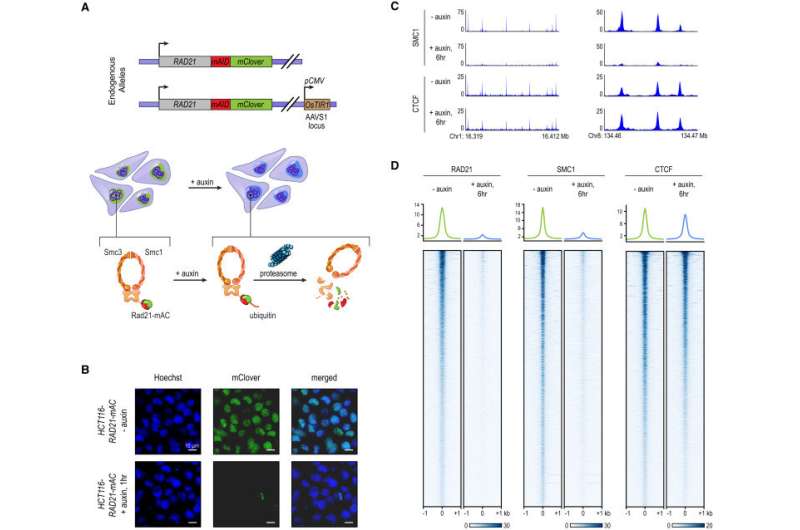
Tagging of Endogenous RAD21 with an Auxin-Inducible Degron Allows for Rapid, Near Complete Cohesin Loss. (A) In HCT-116-RAD21-mAC cells, both RAD21 alleles are tagged with auxin-inducible degrons and an mClover reporter, and the OsTIR1 gene is integrated at the AAVS1 locus. Auxin treatment leads to proteasomal degradation of RAD21. (B) Live cell imaging after Hoechst 33342 staining to label nuclei. Nuclear mClover fluorescence corresponding to tagged RAD21 was lost after 1 hr of auxin treatment. (C) SMC1 and CTCF ChIP-seq signal with and without auxin treatment. (D) RAD21, SMC1, and CTCF ChIP-seq signal (left, middle, right) across all peaks called for each of the proteins in untreated RAD21-mAC cells. Top: Average enrichments for each protein. After RAD21 degradation, the cohesin complex no longer binds to chromatin. CTCF binding is unaffected. Credit: Cell. DOI: 10.1016/j.cell.2017.09.026
A multi-institutional team spanning Baylor College of Medicine, Rice University, Stanford University and the Broad Institute of MIT and Harvard has created the first high-resolution 4-D map of genome folding, tracking an entire human genome as it folds over time. The report, which may lead to new ways of understanding genetic diseases, appears on the cover of Cell.
Making Connections
For decades, researchers have suspected that when a human cell responds to a stimulus, DNA elements that lie far apart in the genome quickly find one another, forming loops along the chromosome. By rearranging these DNA elements in space, the cell is able to change which genes are active.
In 2014, the same team of scientists showed it was possible to map these loops. But the first maps were static, without the ability to watch the loops change. It was unclear whether, in the crowded space of the nucleus, DNA elements could find each other fast enough to control cellular responses.
"Before, we could make maps of how the genome folded when it was in a particular state, but the problem with a static picture is that if nothing ever changes, it's hard to figure out how things work," said Suhas Rao, first author of the new study. "Our current approach is more like making a movie; we can watch folds as they disappear and reappear."
One Ring to Rule them All
To track the folding process over time, the research team began by disrupting cohesin, a ring-shaped protein complex that was located at the boundaries of nearly all known loops. In 2015, the team proposed that cohesin creates DNA loops in the cell nucleus by a process of extrusion.
"Extrusion works like the strap-length adjustor on a backpack," explained Dr. Erez Lieberman Aiden, director of the Center for Genome Architecture at Baylor College of Medicine and senior author on the new study. "When you feed the strap through either side, it forms a loop. DNA seems to be doing the same thing – except that cohesin rings appear to play the role of the adjustor."
Aiden said a crucial prediction of the 2015 model is that all the loops should disappear in the absence of cohesin. In the new research, Aiden, Rao and colleagues tested that assumption.
"We found that when we disrupted cohesin, thousands of loops disappeared," said Rao, a medical student at Stanford University, Hertz Fellow and member of the Aiden lab. "Then, when we put cohesin back, all those loops came back – often in a matter of minutes. This is precisely what you would predict from the extrusion model, and it suggests that the speed at which cohesin moves along DNA is among the fastest for any known human protein."
Loops versus Groups
But not everything happened as the researchers expected. In some cases, loops did the exact opposite of what the researchers anticipated.
"As we watched thousands of loops across the genome get weaker, we noticed a funny pattern," said Aiden, also a McNair Scholar, Hertz Fellow and a senior investigator at Rice University's Center for Theoretical Biological Physics. "There were a few odd loops that were actually becoming stronger. Then, as we put cohesin back, most loops recovered fully – but these odd loops again did the opposite – they disappeared!"
By scrutinizing how the maps changed over time, the team realized that extrusion was not the only mechanism bringing DNA elements together. A second mechanism, called compartmentalization, did not involve cohesin.
"The second mechanism we observed is quite different from extrusion," explained Rao. "Extrusion tends to bring DNA elements together two at a time, and only if they lie on the same chromosome. This other mechanism can connect big groups of elements to one another, even if they lie on different chromosomes. And it seems to be just as fast as extrusion."
Broad Institute Director Eric Lander, a study co-author, said, "We're beginning to understand the rules by which DNA elements come together in the nucleus. Now that we can track the elements as they move over time, the underlying mechanisms are starting to become clearer."
More information: Cohesin Loss Eliminates All Loop Domains. Cell. DOI: dx.doi.org/10.1016/j.cell.2017.09.026 , www.cell.com/cell/fulltext/S0092-8674(17)31120-0
Journal information: Cell
Provided by Baylor College of Medicine