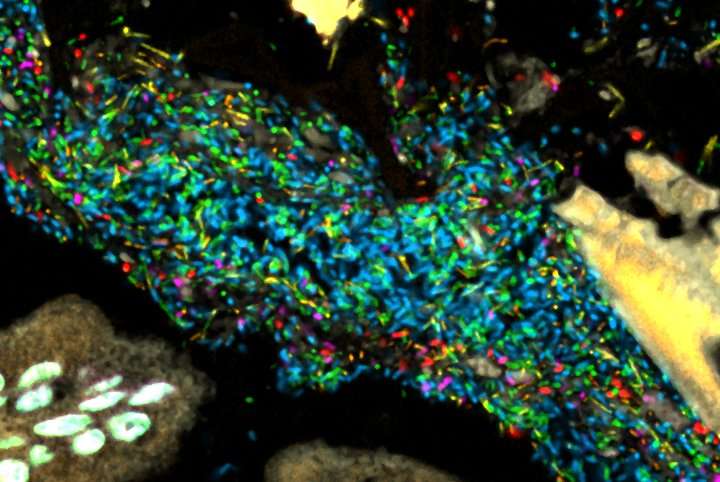
Bacteria in a model human gut microbiome established in a germ-free mouse. Each bacterial species is lit up with a different colored probe, creating a map of the community's spatial organization. Credit: Jessica Mark Welch and Yuko Hasegawa, MBL
Disruptions in the microbiome of the human gut are correlated with several diseases, including obesity and cancer. Yet little is known about the spatial organization of the nearly 1,000 bacterial species in the human gut, which can influence how the species interact with each other and with their host.
In a new collaborative study, scientists from the Marine Biological Laboratory in Woods Hole, the Forsyth Institute, and Washington University in St. Louis established a simplified, model human gut microbiome in germ-free mice and revealed its structure through imaging technologies developed at the MBL. The study is published this week in Proceedings of the National Academy of Sciences.
"We thought we would see clusters of bacteria, with some species congregating around food particles and others abundant in the mucus layer of the gut, which separates the bacteria from host tissue," says MBL scientist Jessica Mark Welch, the lead author of the study. "Instead, we saw a mixed community, where each cell tended to be next to cells of a different species." The bacterial communities near the mucus layer and in the gut's interior (the lumen), where digested food is pushed through by muscular contractions, looked similar.
"The study suggests the host is mixing the microbes and preventing large clusters of single kinds of bacteria from forming," Mark Welch says. "The host does this by sloughing mucus and epithelial cells into the lumen, and by mechanically mixing the contents of the gut. It may be that this mixing creates an evolutionarily stable microbial community."
"No one has looked at a complex microbial community in the gut this way before," says senior author Gary Borisy, a senior research investigator at the Forsyth Institute in Cambridge, Mass. "If we truly want to understand the role of the microbiome, it is not enough to know just which microbes are present. We must also learn what they are doing, who they are talking to and why. Part of the answer to that problem is to figure out who is next to who and who is next to what."
Understanding the spatial structure of microbiomes is a young field that these researchers are pioneering through their novel imaging technology. What they saw in the model gut contrasts with their prior study of human dental plaque, where they discovered highly organized assemblages of bacterial species.
"We don't entirely understand why microbiome organization is so different in the mouth and in the gut," Mark Welch says. "It may have to do with the rate of flow. In the mouth, if bacteria don't adhere to something - either to each other or the host—they end up in the stomach in a matter of seconds. In the gut, flow of contents happens on a timescale of hours rather than seconds."
Mark Welch and collaborators are currently exploring microbiome organization in a number of human and marine ecosystems, including the human tongue, the cuttlefish gut, the surface of kelp, and on marine plastic debris.
Their imaging technology gives the researchers the unique ability to simultaneously image and identify 15 or more microbial taxa, using a technique called Combinatorial Labeling and Spectral Imaging - Fluorescence in situ Hybridization (CLASI-FISH). The team's model gut microbiome contained 15 bacterial species that are typically abundant in the human gut.
More information: Jessica L. Mark Welch et al, Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice, Proceedings of the National Academy of Sciences (2017). DOI: 10.1073/pnas.1711596114
Journal information: Proceedings of the National Academy of Sciences
Provided by Marine Biological Laboratory