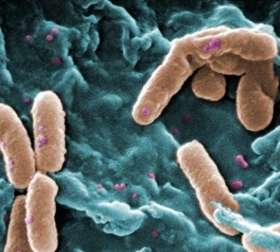
A scanning electron micrograph of Pseudomonas aeruginosa bacteria. Credit: Janice Haney Carr/CDC
A rapid evolutionary "arms race" between bacteria and the killer viruses they contain has been observed by a UNSW-led team of scientists in a sophisticated genetic study of the micro-organisms.
Associate Professor Torsten Thomas, Dr Kerensa McElroy and colleagues from the Centre for Marine Bio-Innovation and the School of Biotechnology and Biomolecular Sciences studied the evolution of Pseudomonas aeruginosa – bacteria that can cause chronic, often lethal lung infections in people with cystic fibrosis.
By sequencing the DNA of entire bacterial populations as they evolved in the laboratory, they were able to identify the genetic mutations underlying rapid evolution of the bacteria.
"We uncovered an interesting arms race between the bacteria and the viruses," says Dr McElroy.
"We found the viruses are evolving unusually rapidly. And the bacteria defend themselves against the viruses by changing their surface structure, so the viruses cannot reinfect them."
The study is published in the journal Proceedings of the National Academy of Sciences.
The research, which makes use of advanced DNA sequencing technologies, is important for understanding how bacteria evolve, so they can be better combated in future.
"Most antibiotics are failing us quite miserably, which has a lot to do with the rapid evolution that bacteria can undergo. It is vital to understand how they evolve, and the different pathways they can take, depending on the environment they live in," says Associate Professor Thomas.
More information: "Strain-specific parallel evolution drives short-term diversification during Pseudomonas aeruginosa biofilm formation." Kerensa E. McElroy, et al. PNAS March 28, 2014. DOI: 10.1073/pnas.1314340111
Journal information: Proceedings of the National Academy of Sciences
Provided by University of New South Wales