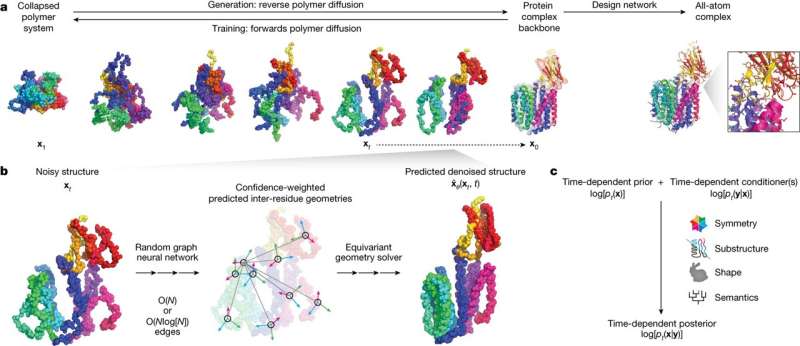
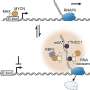
Chroma is a generative model for proteins and protein complexes that combines structured diffusion for protein backbones with scalable molecular neural networks for backbone synthesis and all-atom design. Credit: Nature (2023). DOI: 10.1038/s41586-023-06728-8
A significant step forward in protein design from Generate Biomedicines, Massachusetts, has developed an AI that can generate feasible protein structures and predict the potential functionality of the proteins generated.
In a paper, "Illuminating protein space with a programmable generative model," published in Nature, the team introduces Chroma, a versatile AI capable of producing diverse proteins with specified properties.
Proteins are the amino acid compounds that make up all living things, and these proteins carry out the majority of functions that make life possible. Almost all pharmaceutical drugs target proteins in the body, and many pharmaceuticals are themselves proteins or manufactured using proteins.
With proteins at the core of human health and the pharmaceutical response to disease, being able to create novel proteins to enact specific properties could unlock a cascade of new drug targets. By having a more flexible control over proteins, existing drugs could be made safer and currently untreatable diseases could gain access to previously un-druggable targets.
Chroma's uniqueness lies in its programmability, allowing users to specify a wide array of properties, from inter-residue distances to semantic specifications through classifiers.
The team performed experimental validation tests to challenge Chroma's effectiveness in generating protein designs that express well and exhibit stable folding and structural conformity to the intended design.
Under the most conservative assessment, Chroma exhibited a success rate of approximately 3% in successfully characterized and purified proteins. Chroma showcased proficiency in generating proteins with diverse structures and properties, handling complex shapes, and demonstrating efficient designs, highlighting its potential in tailored protein engineering.
Many untreatable diseases are undruggable, meaning that the protein targets within the body that could be activated or deactivated to counter the disease are too complex or challenging to bind with using existing pharmaceutical proteins.
By back-engineering the properties these targets would require for binding as a starting point for a generative protein assembly, researchers could discover treatments for thousands of diseases. Current efforts are underway to generate arrays of feasible protein structures and then look for potential target matches.
Chroma's capability could shift the focus from generating feasible protein structures towards emphasizing the intended functionality of a protein and forcing the structural formation to follow that intended function.
More information: John B. Ingraham et al, Illuminating protein space with a programmable generative model, Nature (2023). DOI: 10.1038/s41586-023-06728-8
Journal information: Nature
© 2023 Science X Network