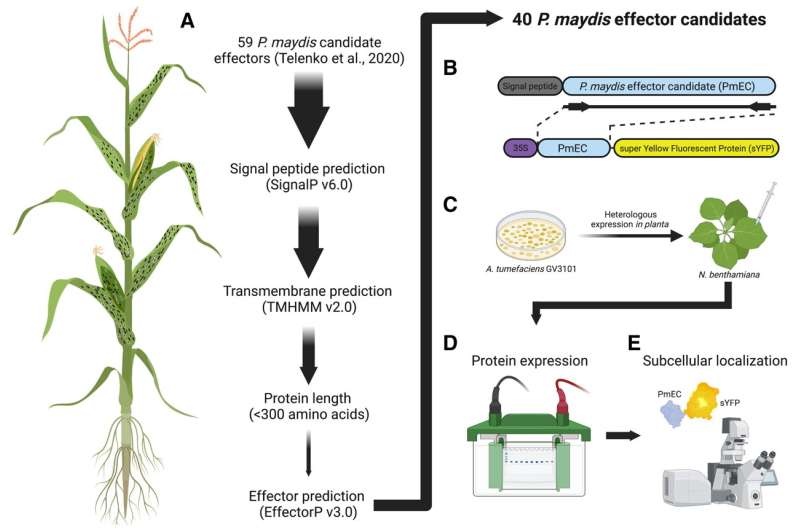
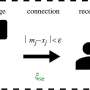
Schematic overview of the selection and subsequent analyses of Phyllachora maydis candidate effectors. A, The P. maydis effector candidates (PmECs) investigated in this study were selected using the aforementioned selection criteria. B, The predicted open reading frames (ORFs) of each of the 40 candidate effectors, without their predicted signal peptides, were synthesized and fused to the N terminus of super yellow fluorescent protein (sYFP) and recombined into the plant expression binary vector pEarleyGate100 (pEG100) using a multisite Gateway cloning strategy. C, The resulting P. maydis effector-fluorescent protein fusion (PmEC:sYFP) constructs were inserted into Agrobacterium tumefaciens for subsequent Nicotiana benthamiana-based heterologous expression assays. D, Immunoblot analyses were used to assess expression of the PmEC-fluorescent protein fusions. E, Laser-scanning confocal microscopy was used to assess the live-cell subcellular localization patterns in N. benthamiana epidermal cells. Figure was created with Biorender. Credit: Phytopathology (2022). DOI: 10.1094/PHYTO-05-22-0181-R
Although discovered in the United States only seven years ago, tar spot has wreaked havoc on corn yield—resulting in an estimated 1.2-billion-dollar loss in 2021 alone. The miscreant behind this devastating plant disease, Phyllachora maydis, is an emergent fungal pathogen whose biology remains obscure. This lack of understanding significantly limits disease management strategies, and no corn germplasm is completely resistant to the pathogen.
Consequently, Dr. Matthew Helm, a Research Molecular Biologist with the Crop Production and Pest Control Research Unit in the USDA-ARS and early career scientists from Purdue University conducted a study to better understand how P. maydis infects corn. Their research, newly published in Phytopathology, is the first publication to characterize this pathogen on a molecular level.
While most fungal pathogens inject plant cells with specialized molecules to suppress host immune responses, the authors investigated whether P. maydis also utilizes this method—in addition to which plant organelles the molecules target. Their data confirms that the tar spot pathogen does encode these virulence molecules and that some of them localize to specific subcellular compartments within the plant cell, including the nucleus and chloroplasts.
The exciting novelty of this study is important, as no other entity has investigated which plant organelles are targeted by pathogen-injected proteins from P. maydis, to the knowledge of Corresponding and First Author Helm. This research will likely impact molecular plant pathology and its subdisciplines significantly.
Helm comments, "Arguably, plant pathologists are only beginning to understand how plant pathogens cause disease on a molecular and genetic level, especially for pathogens that have recently emerged. Our work not only advances our understanding of the biology of this fungal pathogen, but also contributes to our overall understanding of the interaction between plants and microbes."
This study provides the kernel for understanding how this plant disease infects corn—sowing the fields for further studies, increased disease control strategies, and for food security protection worldwide.
More information: Matthew Helm et al, Candidate Effector Proteins from the Maize Tar Spot Pathogen Phyllachora maydis Localize to Diverse Plant Cell Compartments, Phytopathology® (2022). DOI: 10.1094/PHYTO-05-22-0181-R
Provided by American Phytopathological Society