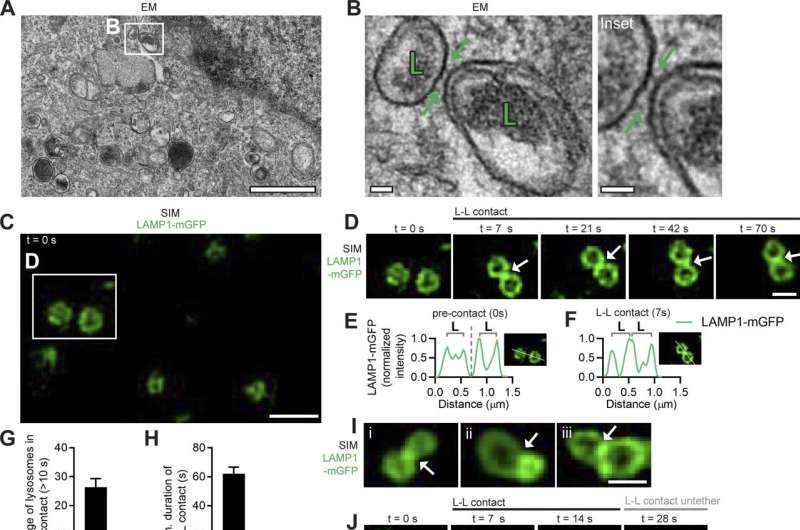
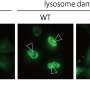
Inter-lysosomal tethering modulates lysosomal network dynamics. (A and B) TEM of two lysosomes tethered together (L, arrows) in untreated HeLa cells. Inset (B) shows inter-lysosomal tether. Scale bars, 1 μm (A); 50 nm (B). (C–F) Super-resolution SIM of inter-lysosomal (L-L) tethering (white arrows, D) in live HeLa cells (LAMP1-mGFP). Inset (D) shows inter-lysosomal tether formation. Corresponding linescans before tether formation (precontact; t = 0 s) and subsequent tethering (contact; t = 7 s) are shown in E and F. Scale bars, 1 μm (C); 0.5 μm (D). Video 1 corresponds to D. (G) Quantification of percentage of lysosomes in an inter-lysosomal tether (duration >10 s) from confocal live-cell microscopy videos (n = 25 cells). (H) Quantification of minimum duration of inter-lysosomal tethering (n = 88 events from 25 cells). (I) Examples of SIM imaging of L-L tethers (white arrows) in live HeLa cells (LAMP1-mGFP). Scale bar, 0.5 μm. (J) SIM imaging of L-L tethering (white arrows) and subsequent L-L contact untethering (yellow arrow) in live HeLa cells (LAMP1-mGFP). Scale bar, 0.5 μm. (K) Confocal microscopy image of lysosomes in live HeLa cells (LAMP1-mGFP) showing inset corresponding to O (t = 0 s). Scale bar, 5 μm. (L) The majority of inter-lysosomal tethers undergo untethering events rather than fusion within 120 s of initial contact formation (n = 64 events from 25 cells). (M and N) Rate of lysosomal untethering events vs. fusion events in live HeLa cells in events/min (M) and frequency of events over time of lysosomal untethering events vs. fusion events (%; N; n = 149 total events from 14 cells). (O–S) Confocal time-lapse microscopy of L-L tethering (white arrows) and subsequent untethering (yellow arrows) in live HeLa cells (LAMP1-mGFP). Scale bars, 0.5 μm. Credit: Journal of Cell Biology (2022). DOI: 10.1083/jcb.202206140
Investigators have discovered that outer mitochondrial membrane proteins regulate crosstalk between mitochondria and lysosomes, according to a Northwestern Medicine study published in the Journal of Cell Biology. These findings have implications for the role of organelle networks in cellular homeostasis and the development of neurological diseases.
"This study further elucidates the mechanism underlying mitochondrial and lysosomal crosstalk and shows how different mutations in the same mitochondrial protein can result in distinct downstream defects in lysosomal network dynamics that contribute to different neurological disorders," said Dimitri Krainc, MD, the Aaron Montgomery Ward Professor and chairman of the Ken and Ruth Davee Department of Neurology.
Lysosomes are organelles tasked with breaking down excess or unusable parts of the cell. Elucidating the regulation of lysosomal networks is key to understanding of cellular dynamics and the role of lysosomes in disease pathogenesis.
In a prior work published in Nature, Krainc's team discovered direct contacts between mitochondria and lysosomes. In the current study, the investigators used live super-resolution microscopy to discover that lysosomes frequently tether together into lysosomal clusters at inter-lysosomal contact sites—sites which modulate lysosomal distribution and function—and subsequently untether, rather than fuse together.
They also found that mitochondria promote this untethering, which is influenced by the hydrolysis of the enzyme Rab7-GTP at inter-lysosomal contact sites. Additionally, they showed that mitochondrial proteins Mid51 and Fis1 form an oligomeric protein complex on mitochondria that, in turn, drives Rab7-GTP hydrolysis and the untethering of lysosomes.
Overall, the study demonstrates how different mutations in Mid51 that are linked to specific neurological disorders result in distinct downstream defects in lysosomal network dynamics.
"A dominant optic atrophy-associated Mid51 mutant which does not disrupt its oligomerization does not disrupt lysosomal network dynamics. In contrast, a Mid51 mutant potentially linked to Parkinson's disease which misregulates its oligomerization disrupts this pathway, resulting in defective lysosomal network dynamics," said Yvette Wong, Ph.D., assistant professor in the Ken and Ruth Davee Department of Neurology's Division of Movement Disorders and lead author of the study.
More information: Yvette C. Wong et al, Mid51/Fis1 mitochondrial oligomerization complex drives lysosomal untethering and network dynamics, Journal of Cell Biology (2022). DOI: 10.1083/jcb.202206140
Journal information: Journal of Cell Biology , Nature
Provided by Northwestern University