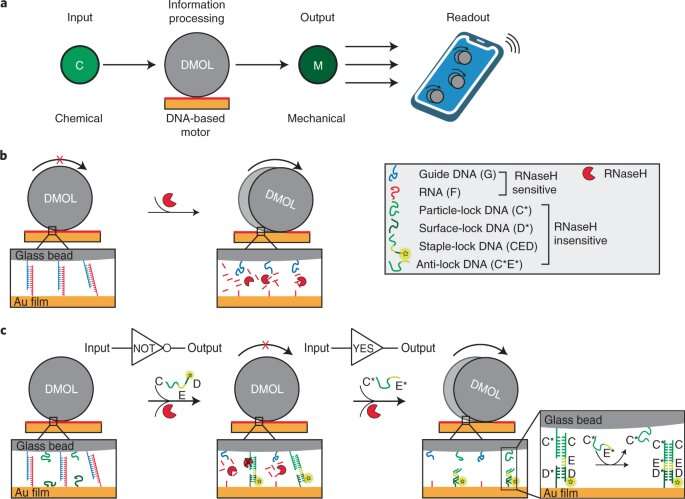
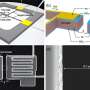
Schematic of DMOLs detecting the presence of a chemical input. Credit: Nature Nanotechnology (2022). DOI: 10.1038/s41565-022-01080-w
A trio of researchers at Emory University has found a way to speed up parallel processing in a DNA computer. In their paper published in the journal Nature Nanotechnology, Selma Piranej, Alisina Bazrafshan and Khalid Salaita describe how they applied DNA as a coating on glass beads and used the results as a type of DNA computer.
A DNA computer is a type of molecular computer that does its work biochemically rather than electronically, as is the case with traditional computers. DNA computers use enzymes that react in certain ways with strands of DNA, which leads to chain reactions. Research involving the use of DNA computers, like quantum computers, has revolved around developing machines that compute in ways not possible on traditional systems.
In this new effort, the researchers took a new approach to creating a DNA computer, applying DNA as a coating to extremely small glass beads. In practice, the glass beads either roll across the surface of a base of gold or hold steady, depending on how the DNA coating interacts with molecules that have been attached to the surface of the chip. In their scenario, a "1" is represented by a roll, while a "0" is represented by a motionless bead. To read the data, the researchers created a smartphone app that analyzes camera imagery. The researchers claim that their DNA computer can provide results in just 15 minutes.
For the computers to work as desired, the researchers included guide DNA molecules in the coating. The strands were engineered to allow for matching RNA molecules on the surface of the chip, preventing a given bead from rolling. But when Ribonuclease H is added to the chip, the interactions with the DNA strands allow for rolling due to breakage of the DNA to RNA complex. The researchers note that their simple computer was able to perform the Boolean operations AND, OR, NOT and YES.
The researchers then added what they describe as a lock complex to control the processing. They note that because of the possibility of using beads with different shapes and sizes, their DNA computer could conceivably output thousands of read-outs in parallel. They suggest one application would be to detect viruses in human saliva samples.
More information: Selma Piranej et al, Chemical-to-mechanical molecular computation using DNA-based motors with onboard logic, Nature Nanotechnology (2022). DOI: 10.1038/s41565-022-01080-w
Journal information: Nature Nanotechnology
© 2022 Science X Network