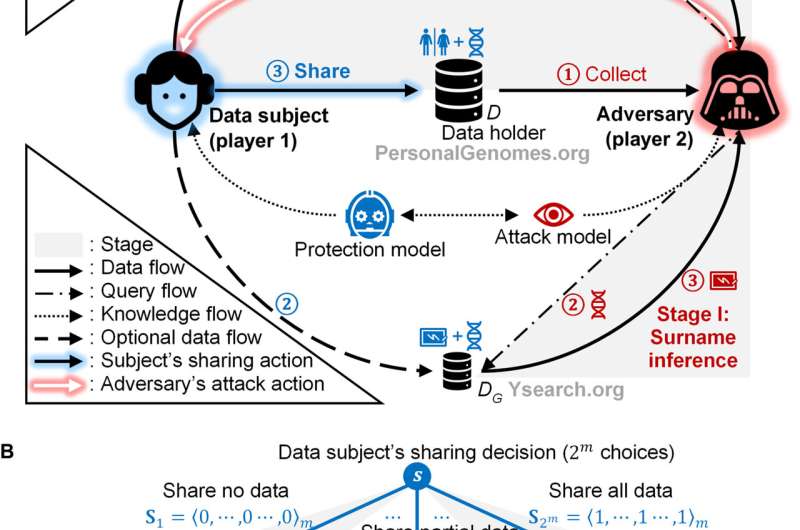
A multistage privacy attack and its game theoretic protection. (A) A system-wide perspective of a multistage re-identification attack and its protection. Person-specific data records of a subject are accessible to an adversary through three databases: a targeted genomic database (D), a genetic genealogy database (DG), and a public identified database (DI). The adversary re-identifies a genomic record by inferring surnames in stage I and linking it to a public record in stage II. The data subject selects a sharing strategy based on a game model only when sharing data in D. (B) A masking game represented in the extensive form. In a masking game, the data subject moves first, and the adversary moves next. Each terminal node is associated with both players’ payoffs. Sj is an m-dimensional vector of 0 and 1 values, representing the jth concrete action of the data subject. More denotation details are in the main text. The opt-in game is a special variation of the masking game in which the data subject only has those two strategies. Credit: Science Advances, 10.1126/sciadv.abe9986
Biomedical data is widely collected in the field of medicine, although sharing such data can raise privacy concerns about the re-identification of seemingly anonymous records. Risk assessment frameworks for formal re-identification can inform decisions on the process of sharing data, and current methods focus on scenarios where data recipients use only one resource to identify purposes. However, this can affect privacy where adversaries can access multiple resources to enhance the chance of their success. In a new report now in Science Advances, Zhiyu Wan and a team of scientists in electrical engineering and computer engineering and biomedical informatics in the U.S. represented a re-identification game using a two-player Stackelberg game of perfect information to assess risk. They suggest an optimal data-sharing strategy based on a privacy-utility trade-off. The team used experiments with large-scale genomic datasets and game theoretic models to induce adversarial capabilities to effectively share data with low re-identification risk.
A new method for data collection
Researchers typically collect most biomedical data on a large scale across a wide range of settings, where personal health data is routinely clinically stored as electronical health records. Biomedical researchers now support studies that collect data across a diverse array of participants and most recent improvements include a number of ventures, including direct-to-consumer genetic testing companies that collect data from various consumers to build repositories. It is assumed that sharing this data beyond their initial point of collection is critical to maximize its social value. However, privacy concerns surrounding such practices include the identifiability of data subjects, including the individuals to whom the data correspond. Genome data are shared across various settings in the United States to provide a clear illustration of the threat of data re-identification and concerns on the possibility. Linking genomic data to identifiers pose a threat to the anonymity of data subjects. In this work, Wan et al. introduced a new method to assess and strategically mitigate risks by explicitly modeling and quantifying the privacy trade-off for subjects during multistage attacks. In this way, the team bridged the gap between more complex models and informed data sharing decisions.
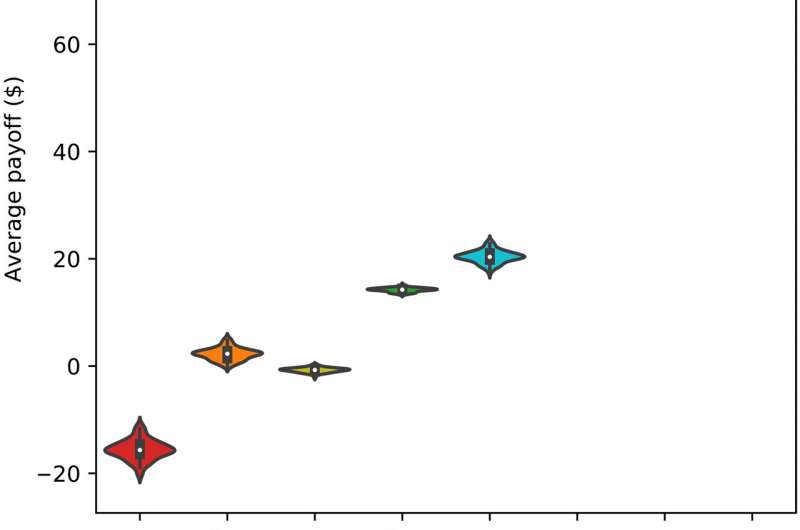
Effectiveness measures of the protection in eight scenarios against a multistage re-identification attack targeting data from 1000 subjects. (A) Violin plot of eight distributions of the data subjects’ average payoffs, where each distribution corresponds to one scenario. The violin plot (depicted using Seaborn) combines boxplot and kernel density estimate for showing the distribution of data subjects’ payoffs in each scenario. A Gaussian kernel is used with default parameter settings. (B) Scatterplot of data subjects’ average privacy metrics and average utility metrics, where each mark corresponds to one scenario and 1 run (of 100 runs). Credit: Science Advances, 10.1126/sciadv.abe9986
Maintaining the privacy of genomic data
Cyberscentists have developed many methods to prevent the re-identification of biomedical data from regulatory and technical perspectives. Nevertheless, most approaches focus on worst-case scenarios, which lead to overestimate the privacy risk. To avert this problem, researchers have introduced risk assessment and mitigation based on game theoretic models. Wan et al. showed how a game theoretic model could reveal the optimal sharing strategy to data subjects in which the team conducted experiments using protection against a multistage attack using either real-world data or large-scale simulations. The results showed how the game theoretic model could efficiently assess and effectively mitigate privacy risks. The sharing strategy or model recommended here could minimize the chance of successfully re-identifying a data subject while maximizing the use of data to keep the released data set useful and the process of data sharing fair.
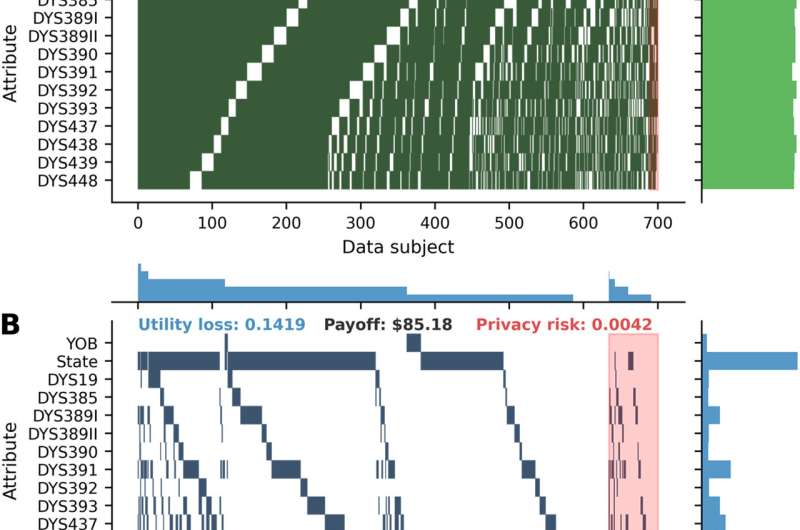
Best strategies for the first 700 data subjects in the first run of experiments in the random masking scenario and three masking game scenarios. (A) Random masking scenario. (B) Masking game. (C) No-attack masking game. (D) One-stage masking game. Each nonwhite block indicates that a data subject masks a specific attribute. Each row represents an attribute, and each column represents a data subject. The distribution for data subjects (attributes) is summarized in the histogram on the top (on the right) with the number of bins equal to the number of data subjects (attributes). Data subjects are split into two groups: those on the left that will not be attacked and those on the right that will be attacked (shaded in red). Columns (or data subjects) within each group are sorted by the number of masked attributes in descending order. Rows (or attributes) are sorted by the order of attributes in the dataset. For each scenario, the average payoff, utility loss, and privacy risk are presented at the top center, top left corner, and top right corner, respectively. For each data subject, utility loss is defined as one minus data utility, and privacy risk is defined as one minus privacy. YOB, year of birth; DYS, DNA Y-chromosome segment. Credit: Science Advances, 10.1126/sciadv.abe9986
Experiments with game theory models.
Wan et al. showed how a game theory model could reveal the optimal sharing strategy by conducting experiments using protection against a multistage attack with either real-world datasets or large-scale simulations, where the game theoretic model could efficiently assess and mitigate the privacy risks. The sharing strategy/model minimized the chance of successfully re-identifying a data subject, while maximizing the use of the data to keep the released dataset useful and the process of data sharing fair. During the experiments, the scientists identified a situation where a data subject could choose how much of their genomic data would be shared in a public repository such as the 1000 Genomes project or the Personal Genome Project. In the scenario, the subject could be willing to share the entire sequence, a subset of short tandem repeats or nothing at all. The goal of this work was to assess the optimal sharing decision of the subject (the leader) relative to the monetary benefit of data sharing and the risk of re-identification by a follower (the adversary). In this model, the subject acted as a leader choosing how much of their genomic data to share and the follower/adversary obtained the shared data to then decide if to execute an attack. When the subject decided and thereby chose a masking strategy, the adversary observed the data with motive depending on the masking strategy.
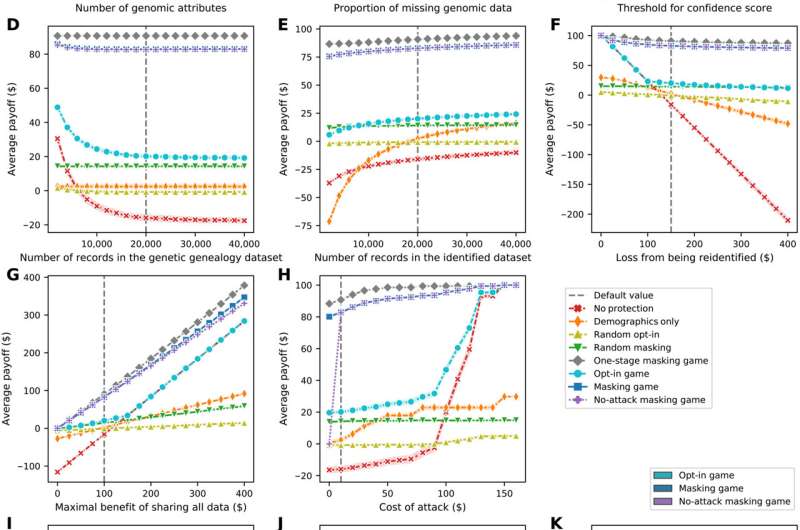
Sensitivity of the data subjects’ average payoff as a function of the parameters and settings in the model. (A) Line plot of the sensitivity to the number of genomic attributes. (B) Line plot of the sensitivity to the proportion of missing genomic data. (C) Line plot of the sensitivity to the threshold for confidence score. (D) Line plot of the sensitivity to the number of records in the genetic genealogy dataset. (E) Line plot of the sensitivity to the number of records in the identified dataset. (F) Line plot of the sensitivity to the loss from being re-identified. (G) Line plot of the sensitivity to the maximal benefit of sharing all data. (H) Line plot of the sensitivity to the cost of an attack. (I) Violin plot of payoff distribution’s sensitivity to the strategy adoption setting. (J) Violin plot of the sensitivity to the surname inference approach. (K) Violin plot of the sensitivity to the weight distribution of attributes. Each line plot (depicted using Seaborn) shows data subjects’ average payoffs, with error bars representing SDs, in eight scenarios. Each violin plot (depicted using Seaborn) combines boxplot and kernel density estimate for showing the distributions of data subjects’ average payoffs in several scenarios. Gaussian kernels are used with default parameter settings. TMRCA, time to most recent common ancestor; KNN, k-nearest neighbors. Credit: Science Advances, 10.1126/sciadv.abe9986
Outlook: Data analysis and anonymization
The team then simulated populations that maintained 20 attributes including ID and surname, as well as 16 genomic characteristics. The simulation compared several scenarios in which the adversary claimed to re-identify all records in all scenarios. Among them, the 'no-protection' scenario showed the highest variation relative to data utility. The team used a machine with a six-core, 64-bit central processing unit to compute the resulting strategies for all 1000 data subjects in each scenario.
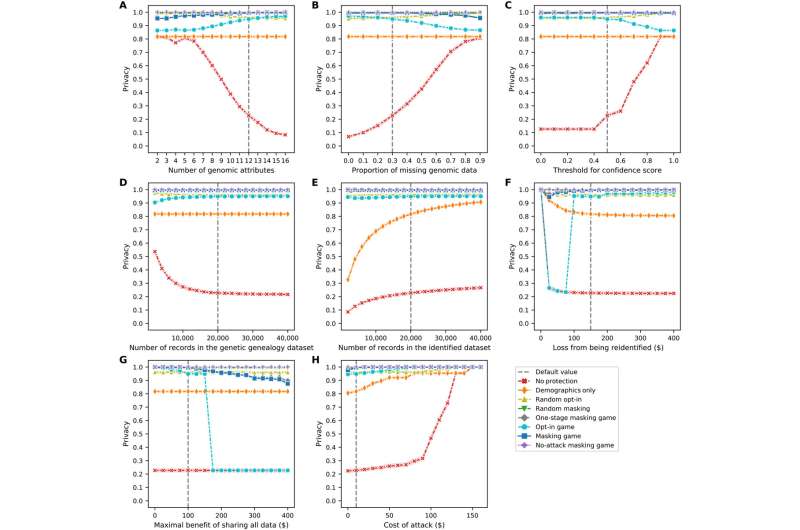
Sensitivity of the data subjects’ average privacy as a function of the parameters in the model. (A) Number of genomic attributes. (B) Proportion of missing genomic data. (C) Threshold for confidence score. (D) Number of records in the genetic genealogy dataset. (E) Number of records in the identified dataset. (F) Loss from being re-identified. (G) Maximal benefit of sharing all data. (H) Cost of an attack. Each line plot (depicted using Seaborn) shows the data subjects’ average payoffs, with error bars representing SDs, in eight scenarios. Credit: Science Advances, 10.1126/sciadv.abe9986
Wan et al. also tested the sensitivity of the model against eight parameters and three experimental settings and conducted a cast study to show the applications of the model to real-world datasets. Using Craig Venter's demographic attributes including year of birth, state of residence, and gender, the method allowed the subjects to make informed data-sharing decisions when faced with complex, state-of-the-art re-identification models. The flexible method can help answer questions on the risks of sharing de-identified data to an open data repository, and help individuals discern the portion of the data to be shared. In this way, Zhiyu Wan and colleagues discuss the game theoretic model, including its limitations and note how it can be expanded to provide directions for future work. For instance, the team envisions integrating the solution as a service into existing anonymization software.
More information: 1. Zhiyu Wan et al, Using game theory to thwart multistage privacy intrusions when sharing data, Science Advances (2021). DOI: 10.1126/sciadv.abe9986
2. W. Nicholson Price et al, Privacy in the age of medical big data, Nature Medicine (2018). DOI: 10.1038/s41591-018-0272-7
Journal information: Science Advances , Nature Medicine
© 2021 Science X Network