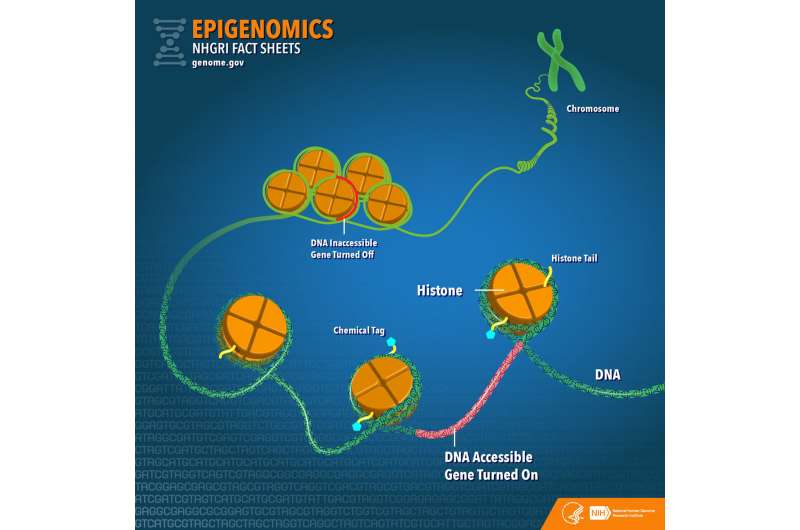
An artistic representation of gene regulating elements, which allow cells with the same genetic code to differentiate into many different tissues and play many varied roles in the body. Credit: Ella Maru Studio
A 17-year research project has generated a detailed atlas of the genome that reveals the location of hundreds of thousands of potential regulatory regions—a resource that will help all human biology research moving forward.
Of the three billion base pairs in the human genome, only 2% code for the proteins that build and maintain our bodies. The other 98% harbors, among other things, potential regulatory regions—sequences that give cells the instructions and tools needed to turn protein recipes into an astonishingly complex organism. Yet despite their importance and prevalence, non-coding regions have been studied much less than gene-coding sequences, in part because it is more difficult to do so.
The Encyclopedia of DNA Elements (ENCODE) collaboration was launched by the National Human Genome Research Institute with the goal of developing the tools and expertise needed to shed light on our genome's mysterious majority. Now in its final year, ENCODE has made huge advances thanks to the combined scientific and technological prowess of several hundred researchers at dozens of institutions.
"We've sequenced the human genome and we largely know where genes are. But when you get outside genes, mapping the function of genomic 'dark matter' is much more daunting. It's a big step forward for us to know how to find the areas within the 98% that are functionally important," said Len Pennacchio, a senior scientist at Lawrence Berkeley National Laboratory (Berkeley Lab) and co-author on four of the 15 new ENCODE papers published this week as part of a special collection in Nature. In addition to their original research, Pennacchio and his Berkeley Lab colleagues also provided technical expertise and materials to other ENCODE consortium teams.
An illustration of DNA modifying elements, including histones and chemical tags. Credit: Lawrence Berkeley National Laboratory
According to Pennacchio, the project's recent advances will be particularly useful for scientists studying diseases. When trying to determine the underlying causes of a condition, researchers search for genetic variants carried by affected individuals. Sometimes, he said, they find associations with sequences within genes, but often the analyses will pinpoint an area that's far away from any protein-coding sequence, and it isn't readily apparent what that DNA does. Is it important in the heart, or the stomach? Is it important all the time or just at certain phases of development?
"Our datasets give scientists clues as to when and where that sequence functions, and which gene or genes it affects. It gives you an immediate path to follow to learn more, where previously we'd have few hints," he said.
More information: Chung-Chau Hon et al. Expanded ENCODE delivers invaluable genomic encyclopedia, Nature (2020). DOI: 10.1038/d41586-020-02139-1
Journal information: Nature
Provided by Lawrence Berkeley National Laboratory