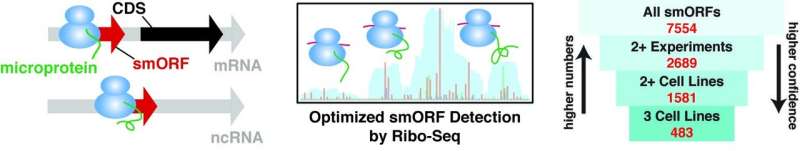
This illustration represents the Saghatelian lab's method for finding genes known as small open reading frames (smORFs). The "microproteins" encoded by smORFs have been linked to immune function, cell stress and many other cellular processes, which suggests that detecting smORFs could lead scientists to new biomarkers and drug targets for human diseases. Credit: Salk Institute
While scientists know of about 25,000 genes that code for biologically important proteins, additional, smaller genes hiding in our DNA may be just as important. But these tiny lines of genetic code have proven tough to track down.
A new study from the Salk Institute identified over 2,000 new, small genes—expanding the number of human genes by 10 percent. These previously unknown genes are known as small open reading frames (smORFs), and the scientists have developed a method for detecting these important genetic sequences in human cell lines.
"We've expanded the human genome," says Salk Professor Alan Saghatelian, co-corresponding author of the study, published in Nature Chemical Biology on December 9, 2019. "This work can really be applied to better understand human biology and may eventually have implications for diseases ranging from cancer to diabetes."
Over the last ten years, Saghatelian and his colleagues have been developing methods to better identify smORFs that affect human health. Already, "microproteins" encoded by smORFs have been linked to immune function, cell stress and even early muscle development. Saghatelian says there is growing evidence that detecting smORFs could lead scientists to new biomarkers and drug targets for human diseases.
Thomas Martinez, first author of the study and postdoctoral fellow in the Saghatelian lab, led the effort to use a technique called Ribo-Seq to see which smORFS actually encoded proteins in cells. Ribo-Seq is routinely used for detecting the production of larger proteins but proved less consistent for detecting smORFs. The team solved this problem by optimizing the experiment to more reliably detect smORFs and yield the most robust estimate of the number smORFs in the human genome.
From left: Alan Saghatelian and Thomas Martinez Credit: Salk Institute
Martinez's work made it possible to find smORFs in three human cell lines, taken from leukemia, ovarian cancer and immortalized kidney cells. Around 7,500 smORFs showed up in at least one cell line. Of those, around 1,500 appeared in at least two cell lines—and kept showing up when the researchers repeated their experiments. The reproducibility of the results gave the researchers confidence that these newly spotted genes really existed.
"We finally have reliable information that the human genome contains at least 2,500 to 3,500 smORFs," says Saghatelian.
The challenge now is to figure out which smORFs are involved in disease—and whether the microproteins they code for could be disease targets. Already, the researchers have identified around 500 smORFs that show up in all three cell lines, suggesting they could have important biological functions.
"Right now, our methods can tell us if a smORF exists or doesn't exist, but it doesn't give us a lot of information on what is actually related to disease," says Saghatelian. "Going forward, the lab will start doing more research to find smORFs that may be specific to diseases like cancer or diabetes."
Saghatelian says the science of smORFs is still in its early days, so the researchers hope other labs around the world will use their methods to hunt for smORFs in their own cell lines.
"This is really an unexplored area," says Martinez. "At the end of the day, you want to know what all the parts are in the genome."
More information: Accurate annotation of human protein-coding small open reading frames, Nature Chemical Biology (2019). DOI: 10.1038/s41589-019-0425-0 , nature.com/articles/s41589-019-0425-0
Journal information: Nature Chemical Biology
Provided by Salk Institute