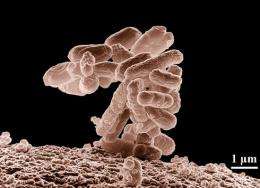
Electron micrograph image of a cluster of E. coli bacteria magnified 10,000 times. Researchers were able to quickly sequence the DNA of a suspected strain of E. coli thought to be the case of the latest outbreak in Europe. Credit: Eric Erbe | Christopher Pooley | USDA | ARS | EMU
The discovery of how a group of bacteria rapidly adapts to changing growth conditions could have implications for future antibiotic development, according to research at the University of Oxford and the University of York.
Led by Professor Colin Kleanthous at Oxford and Dr Christoph Baumann at York, the research which also involved key collaborators Mark Sansom at Oxford and Jacob Piehler at the University of Osnabrück, is published in Nature.
Gram-negative bacteria are a major cause of disease, in part because they have a robust outer membrane that protects against the immune system and certain antibiotics. They can live in a broad range of environments, which for E. coli includes river water as well as humans and animals.
The bacteria have intricate regulatory mechanisms for ensuring they have the right complement of outer membrane proteins - known as OMPs - for a particular habitat. But little is known about how OMPs are replaced in the outer membrane when bacteria adapt to changes in their growth conditions.
The new research describes how bacteria are able to change the proteins in their outer membrane and how this is intimately linked to the process of protein insertion in the membrane.
The researchers tracked how colicins—toxins produced by some strains of E. coli—make their way into bacteria via specific OMP receptors. Using single-molecule fluorescence microscopy, they noticed that the colicin-bound receptors behaved in an unusual way in the membrane.
"We spent many years trying to figure out what might be causing the receptors to behave in this way, as if something was boxing them in," Professor Kleanthous explained.
The researchers discovered that hundreds of the receptors bunch together in the outer membrane into structures they call 'OMP islands' causing the 'boxing in' effect.
The scientists replicated the apparently complex behaviour of OMPs in Gram-negative bacteria using purified proteins in an artificial membrane system in which they found that OMPs have a natural tendency to self-associate.
OMP islands contain an important molecular machine that is responsible for the insertion of new OMPs in the membrane. "We were not surprised to find the OMP insertion machinery in the islands, but it was completely unexpected to discover this machinery shuts down as the outer membrane matures," Dr Baumann, of the Department of Biology at York, explained. Although the reason for this is unclear, it is an important part of the new mechanism as it means 'old' and 'new' OMPs do not intermix.
The researchers found that old OMPs are pushed to the ends of a growing cell as new OMPs are inserted in the central region of the cell. After two cell divisions, cells appear that do not have any of the original OMPs. In simple terms, a bacterium like E. coli can change its outer membrane protein coat in just two generations.
Professor Kleanthous and Dr Baumann believe that their ongoing collaborative work, which began when the former was a member of the Department of Biology at York, will have a major impact on our understanding of the bacterial outer membrane.
"It offers up many new avenues of research and also suggests new potential targets for antibiotic development, for example the disruption of OMP islands," they said.
More information: 'Supramolecular assemblies underpin turnover of outer membrane proteins in bacteria. Rassam P, Copeland NA, Birkholz O, Toth C, Chavent M, Duncan A, Cross SJ, Housden NG, Kaminska R, Seger U, Quinn DM, Garrod TJ, Sansom MSP, Piehler J, Baumann CG and Kleanthous C. Nature (June 2015) DOI: 10.1038/nature14461
Journal information: Nature
Provided by University of York