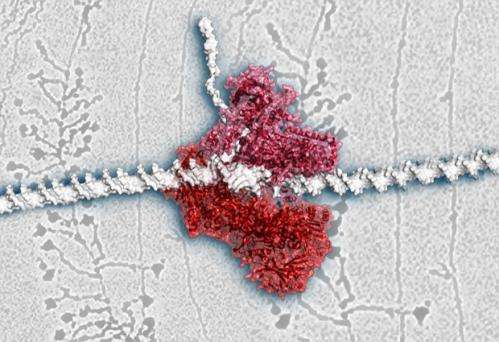
This is the structure of RNA polymerase I revealed an efficiency-boosting strategy. Credit: EMBL/C. Férnandez-Tornero & P. Riedinger
The molecular machine that makes essential components of ribosomes – the cell's protein factories – is like a Swiss-army knife, researchers at the European Molecular Biology Laboratory (EMBL) in Heidelberg, Germany, and the Centro de Investigaciones Biológicas in Madrid, Spain, have found. By determining the 3-dimensional structure of this machine, called RNA polymerase I, for the first time, the scientists found that it incorporates modules which prevent it from having to recruit outside help. The findings, published online today in Nature, can help explain why this protein works faster than its better-studied counterpart, RNA polymerase II.
"Rather than recruiting certain components from outside, RNA polymerase I has them already built in, which explains why it is bigger, and less regulated, but at the same time more efficient," says Christoph Müller from EMBL, who led the study. "Because everything is already assembled, there's no time delay," explains Maria Moreno-Morcillo, who carried out the work.
There are three different RNA polymerases, each of which makes specific types of RNA molecule. For example, RNA polymerase II makes messenger RNA – the 'middle-man' that carries the information encoded in DNA to a ribosome where it can be used to make a protein. RNA polymerases I and III make parts of the machinery which reads that messenger RNA: I builds the RNA that will eventually form a ribosome, while III makes the transfer RNA that carries the protein building blocks to the ribosome for assembly. Scientists have known for over a decade what RNA polymerase II looks like and how it works, but obtaining detailed information on the structures of its counterparts has proven extremely difficult. Now that they have managed to do so for RNA polymerase I, Müller and colleagues have found explanations for some of the protein's particularities.
Part of the difficulty in studying RNA polymerase I is that it is a larger molecule than RNA polymerase II. When they determined its 3-dimensional structure, the scientists found that some of the 'extra' modules in RNA polymerase I are remarkably similar to other, separate proteins that RNA polymerase II needs to do its job. It seems that RNA polymerase I has brought those helper modules permanently on board. In another part of the molecule, Müller and colleagues found that RNA polymerase I appears to have combined what in RNA polymerase II are two separate modules into a single, multi-tasking component. Together, these changes likely explain why RNA polymerase I can produce RNA molecules at a faster rate than RNA polymerase II.
The findings also imply that the cell has fewer ways of controlling RNA polymerase I's activity, since it can't influence it by changing the availability of helper proteins as it does in the case of RNA polymerase II. But here, too, RNA polymerase I's Swiss-army knife strategy provides a solution. The structure showed that this molecular machine has a built-in regulatory mechanism: it can stop itself from attaching to DNA by bending a loop in its structure to block the space the DNA would usually dock onto.
More information: Fernández-Tornero, C., Moreno-Morcillo, M., Rashid, U.J., Taylor, N.M.I, Ruiz, F.M., Gruene, T., Legrand, P., Steuerwald, U. & Müller, C.W. Crystal structure of the 14-subunit RNA polymerase I. Published online in Nature on 23 October 2013. DOI: 10.1038/nature12636
Journal information: Nature
Provided by European Molecular Biology Laboratory