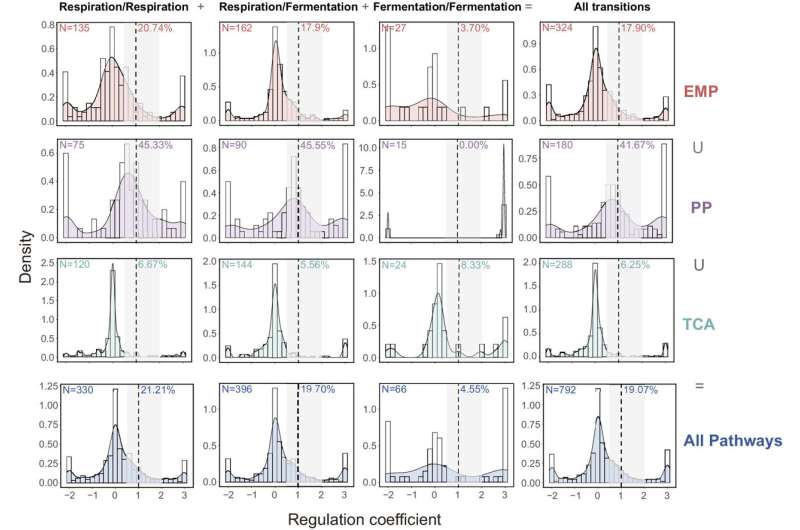
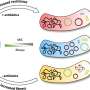
Distributions of coefficients for metabolic concentration regulation ( ρm ). The number of flux changes considered for each pathway is given in the upper left corner (N). The percentages in each plot quantify the fraction of flux changes explained by the corresponding mechanism ( 0.5<ρm<2 , gray interval). Different colors represent different metabolic pathways. Respiration/Respiration means both pairs of transitions were at respirative metabolism. Fermentation/Fermentation means both pairs were at respiro-fermentative metabolism. Respiration/Fermentation means pairs of transitions between respirative and respiro-fermentative metabolism. EMP, Embden–Meyerhof–Parnas pathway; PP, pentose phosphate pathway; TCA, tricarboxylic acid cycle. Credit: Proceedings of the National Academy of Sciences (2023). DOI: 10.1073/pnas.2302779120
Metabolic reaction flux change is the final result of interacting regulations by intracellular gene expression, transcriptional regulation, protein level, translation modification, and allosteric effect. However, the regulatory mechanism of metabolic flux changes is still not very clear.
For the central carbon metabolic pathway, the contribution proportion of two important regulatory factors, protein phosphorylation and the allosteric effects of metabolites, to flux changes has not been quantified, and the metabolic regulatory mechanism behind the Crabtree effect of Saccharomyces cerevisiae is still not fully understood. For example, the regulatory factors that mainly contribute to the metabolic flux changes through the central carbon metabolic pathway under aerobic respiration and aerobic fermentation conditions have not been reported.
In a study published in Proceedings of the National Academy of Sciences, Xia Jianye's team from the Tianjin Institute of Industrial Biotechnology of the Chinese Academy of Sciences, Zhuang Yingping's team from East China University of Science and Technology, and Jens Nielsen's team from the Chalmers University of Technology, developed a method to identify allosteric effectors that have physiological reaction flux regulation functions and quantified the proportion of each regulatory factor (protein abundance, thermodynamic potential, substrate concentration, allosteric effector concentration, and protein phosphorylation) contributing to flux changes.
By 13C isotopically labeled metabolic flux analysis, scientists first obtained flux distribution of the central carbon metabolic pathway of Saccharomyces cerevisiae under aerobic respiration and aerobic fermentation conditions. They used hierarchical regulatory analysis to quantify the contributed proportion of protein abundance and thermodynamic potentials to flux changes.
Then, scientists used the Bayesian inference approach to identify the allosteric effectors that have reaction flux regulation functions. They quantified contributions of these physiologically relevant allosteric effectors combined with the substrates to reaction flux changes.
The protein phosphorylation analysis showed that protein phosphorylation plays a key role in regulating the flux of the glycolytic pathway in Saccharomyces cerevisiae.
This study showed that the regulatory patterns of metabolic flux changes in the central carbon metabolic pathway of Saccharomyces cerevisiae under both aerobic respiration and aerobic fermentation conditions. The method used in this study can be used in the study of metabolic flux regulation mechanisms in other strains.
More information: Min Chen et al, Yeast increases glycolytic flux to support higher growth rates accompanied by decreased metabolite regulation and lower protein phosphorylation, Proceedings of the National Academy of Sciences (2023). DOI: 10.1073/pnas.2302779120
Journal information: Proceedings of the National Academy of Sciences
Provided by Chinese Academy of Sciences