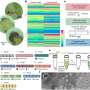
Structure and functional analysis of tRNA-Asp-AUC. (A) Predicted structure of the SPC6 tRNA-Asp-AUC. Solid circles represent G:C base pairs and open circles represent A/G:U base-pairs. (B) Relative eGFP expression in vegetative hyphae of S. coelicolor strains, each containing an integrated eGFP gene with from 0 up to 6 copies of an N-terminal GAT codon (D). Error bars indicate SD, *** signifies P ≤ 0.001. (C) (i) Fluorescence microscopy, differential interference contrast and merged images of vegetative hyphae of SPC6 and S. coelicolor M145 containing the integrated plasmid pIJRCG encoding a modified eGFP with 6 N-terminal aspartic acid residues translated from repeated GAU codons [D], as indicated above the hyphal images. Also shown are hyphae of SPC6 containing the cloning vector pIJ8660 with a promoterless unmodified copy of eGFP. Strains were grown for 3 days at their optimal temperatures (30°C for S. coelicolor M145 and 37°C for SPC6). (ii) Microscopy of a representative aerial hypha of S. coelicolor M145 containing pIJRCG, sampled after 6 days growth. The arrow indicates a spore. Bar = 10 uM. (D) Microscopy of a representative vegetative hypha, sampled after 3 days growth, of S. coelicolor M145 containing the integrated plasmid pIJTCG, in which the SPC6 tRNA-Asp-AUC is co-expressed, as indicated above the hyphal images. Bar = 10 uM. Credit: Nucleic Acids Research (2022). DOI: 10.1093/nar/gkac502
A research team led by Prof. Liu Guangxiu from the Northwest Institute of Eco-Environment and Resources of the Chinese Academy of Sciences (CAS) has isolated a fast-growing desert streptomycete from an extreme environment in the Badain Jaran Desert, and discovered a new tRNA-ASP-AUC gene in its genome.
Related results were published in Nucleic Acids Research.
Streptomycete is a major group of microorganisms used to synthesize natural products and drugs, and its genome contains a large number of secondary metabolic gene clusters.
Although many valuable secondary metabolites, such as streptomycin, natamycin and chloramphenicol, have been identified from a variety of streptomycete species and have been used in human clinical treatment, animal breeding, crop resistance and other fields, these compounds account for less than 5% of the discovered secondary metabolic gene clusters.
At the same time, many existing fermentation processes have defects such as low antibiotic yield and a long cycle. Therefore, how to effectively activate these secondary metabolic gene clusters or improve the fermentation yield of existing antibiotics is key in the study of microbial antibiotics.
In this study, the researchers described the function of a novel tRNA-Asp-AUC found in the SPC6 genome, and demonstrated how its heterologous expression both in the model streptomycete, S. coelicolor, and in a range of Streptomyces species used by the pharmaceutical industry, increased production of antibiotics.
This novel tRNA-Asp-AUC gene can not only effectively identify the rare codon GAT of aspartic acid in streptomycete to promote the expression of secondary metabolic gene clusters, but also regulate the synthesis of vitamin B12 related compounds, which is a key factor in antibiotic synthesis, thus accelerating the synthesis of secondary metabolites and improving their yield.
The study provides an efficient scheme to enhance the rate of antibiotic synthesis in industrial streptomycete, and opens up an effective way to develop new secondary metabolites from steptomycete.
More information: Ximing Chen et al, A new bacterial tRNA enhances antibiotic production in Streptomyces by circumventing inefficient wobble base-pairing, Nucleic Acids Research (2022). DOI: 10.1093/nar/gkac502
Journal information: Nucleic Acids Research
Provided by Chinese Academy of Sciences
![Structure and functional analysis of tRNA-Asp-AUC. (A) Predicted structure of the SPC6 tRNA-Asp-AUC. Solid circles represent G:C base pairs and open circles represent A/G:U base-pairs. (B) Relative eGFP expression in vegetative hyphae of S. coelicolor strains, each containing an integrated eGFP gene with from 0 up to 6 copies of an N-terminal GAT codon (D). Error bars indicate SD, *** signifies P ≤ 0.001. (C) (i) Fluorescence microscopy, differential interference contrast and merged images of vegetative hyphae of SPC6 and S. coelicolor M145 containing the integrated plasmid pIJRCG encoding a modified eGFP with 6 N-terminal aspartic acid residues translated from repeated GAU codons [D], as indicated above the hyphal images. Also shown are hyphae of SPC6 containing the cloning vector pIJ8660 with a promoterless unmodified copy of eGFP. Strains were grown for 3 days at their optimal temperatures (30°C for S. coelicolor M145 and 37°C for SPC6). (ii) Microscopy of a representative aerial hypha of S. coelicolor M145 containing pIJRCG, sampled after 6 days growth. The arrow indicates a spore. Bar = 10 uM. (D) Microscopy of a representative vegetative hypha, sampled after 3 days growth, of S. coelicolor M145 containing the integrated plasmid pIJTCG, in which the SPC6 tRNA-Asp-AUC is co-expressed, as indicated above the hyphal images. Bar = 10 uM. Credit: Nucleic Acids Research (2022). DOI: 10.1093/nar/gkac502 New tRNA promotes synthesis of streptomyces antibiotics](https://scx1.b-cdn.net/csz/news/800a/2022/new-trna-promotes-synt.jpg)