Researchers rethink how our feathered friends evolved
A recently published global genome study that used the data-intensive Gordon supercomputer at the San Diego Supercomputer at the University of California, San Diego, has researchers rethinking how avian lineages diverged after the extinction of the dinosaurs.
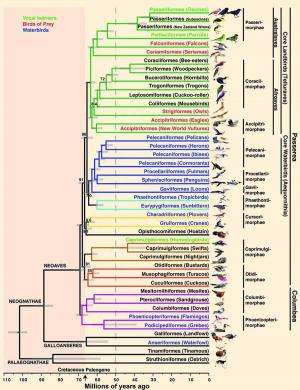
The four-year project, called the Avian Genome Consortium and published in the journal Science, resulted in a new family "tree" for nearly all of the 10,000 species of birds alive today by comparing the entire DNA codes (genomes) of 48 species as varied as parrot, penguin, downy woodpecker, and Anna's hummingbird. The massive undertaking, started in 2011, involved more than 200 researchers at 80 institutions in 20 countries, with related studies involving scientists at more than 140 institutions worldwide.
The genome-scale phylogenetic analysis of the 48 bird species considered approximately 14,000 genes. This presented computational challenges not previously encountered by researchers in smaller-scale phylogenomic studies based on analyses of only a few dozen genes. The inclusion of hundreds of times more genetic data per species allowed the researchers to realize the existence of new inter-avian relationships.
"Characterization of genomic biodiversity through comprehensive species sampling has the potential to change our understanding of evolution," wrote Erich Jarvis, associate professor of neurobiology at the Howard Hughes Medical Institute at Duke University and the study's principal investigator, in an introduction to a special issue of the journal Science containing eight papers from the study. An additional 20 papers generated by the study were simultaneously published in other journals.
"For 50 species, more than 10 to the power of 76 possible trees of life exist. Of these, the right one has to be found," said Andre J. Aberer, with the Heidelberg Institute for Theoretical Studies (HITS), in a news release at the time of the study's publication in Science. "For comparison: About 10 to the power of 78 atoms exist in the universe."
Many of the computations were done on SDSC's Gordon supercomputer by Aberer with the assistance of SDSC Distinguished Scientist Wayne Pfeiffer. They ran a new code called ExaML (Exascale Maximum Likelihood) to infer phylogenetic trees using Gordon soon after it debuted in 2012 as one of the 50 most powerful supercomputers in the world.
Developed by Alexandros Stamatakis, head of the Scientific Computing Group at HITS, ExaML couples the popular RAxML search algorithm for inference of phylogenetic trees using maximum likelihood with an innovative MPI parallelization approach. This yields improved parallel efficiency, especially on partitioned multi-gene or whole-genome data sets.
"I had previously collaborated with Alexis on improving the performance of RAxML," said Pfeiffer. "He described the goals of the Avian Genome Consortium, and we agreed that Gordon, with its just-released fast processors, could provide much of the computer time needed for this ambitious project. In the end, more than 400,000 core hours of computer time were consumed on Gordon."
"After doing initial analyses on our institutional cluster, we rapidly realized that comprehensive analysis of the more challenging data sets being considered would require supercomputer resources," said Aberer. "Access to Gordon was thus invaluable for achieving results in a timely manner."
Discover the latest in science, tech, and space with over 100,000 subscribers who rely on Phys.org for daily insights. Sign up for our free newsletter and get updates on breakthroughs, innovations, and research that matter—daily or weekly.
In all, high-performance computing (HPC) resources at nine supercomputer centers were used to analyze the complete genomes because of the scope of the undertaking. In addition to Gordon, several other U.S.-based supercomputers that are or have been part of the National Science Foundation's eXtreme Science Engineering and Discovery Environment (XSEDE) were used: Ranger, Lonestar, and Stampede at the Texas Advanced Computing Center (TACC) at the University of Texas at Austin; and Nautilus at the National Institute of Computational Sciences (NICS) at the University of Tennessee.
Resolving the timing and phylogenetic relationships of birds is important not only for comparative genomics, but can also inform about human traits and diseases, according to the researchers. For example, the study included vocal-learning species - such as parrots and hummingbirds - which can serve as models for spoken language in humans and may prove useful for insights into speech disorders.
More information: Whole-genome analyses resolve early branches in the tree of life of modern birds, Science 12 December 2014: Vol. 346 no. 6215 pp. 1320-1331 DOI: 10.1126/science.1253451
Journal information: Science
Provided by University of California - San Diego