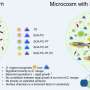
Researchers worked with samples extracted from Norris Geyser Basin at Yellowstone National Park. Credit: Tjflex2, CC BY-NC-NC-2.0
Researchers determined the contributions of different microbes toward the establishment of microbial mat communities in the hot and acidic environments of the Yellowstone Hot Springs.
Microbial mats serve as model systems for studying microbial interactions and their influence over biogeochemical processes. Understanding how communities of microbes establish mats over time provides insights on how their environments determine the mechanisms they employ.
Microbes such as those that thrive in the extreme environments of Yellowstone Hot Springs have been part of studies conducted at the U.S. Department of Energy Joint Genome Institute (DOE JGI), a DOE Office of Science User Facility, for their potential bioenergy and environmental applications. A team of researchers from the Pacific Northwest National Laboratory and DOE JGI, led by longtime DOE JGI collaborator Bill Inskeep of Montana State University, developed a conceptual model that details how microbial mats are formed in hot, acidic springs in the Yellowstone caldera. The team sequenced DNA samples extracted from two acidic geothermal springs at various timepoints over two months in Norris Geyser Basin at Yellowstone National Park.
The data allowed the team to track the formation of microbial mats, beginning with primary colonization by Hydrogenobaculum species and Metallosphaera yellowstonensis, and how these populations as well as those of other microbes that colonized later changed over time in response to availability of nutrients such as oxygen and carbon. These studies continue to build on the decades of microbial field studies Inskeep and his team have done at Yellowstone National Park. The insights gained from this model, the team noted, could provide insights into microbial life at other hot springs ecosystems and, potentially, on other planets.
More information: Beam JP et al. Assembly and Succession of Iron Oxide Microbial Mat Communities in Acidic Geothermal Springs. Front. Microbiol. (2016) DOI: 10.3389/fmicb.2016.00025
Provided by DOE/Joint Genome Institute