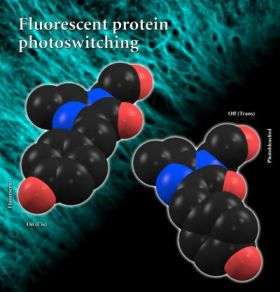
Graphic shows models of the on and off structural alignments of a photoswitchable fluorescent protein. Credit: Courtesy S. James Remington
University of Oregon scientists have identified molecular features that determine the light-emitting ability green fluorescent proteins, and by strategically inserting a single oxygen atom they were able to keep the lights turned off for up to 65 hours.
The findings, published online this week by the Proceedings of the National Academy of Sciences, likely are applicable to most photoswitchable fluorescent proteins, said S. James Remington, professor of physics and member of the UO Institute of Molecular Biology.
"This new model makes specific predictions and improves the qualities of the protein as a photo-switchable label," Remington said. "It gives us the first picture of how these molecules can be switched on and off. That allows us to design new variants to make the proteins more useful."
For more than a decade, fluorescent proteins – first isolated in jellyfish and since found in a variety of colors from coral reef organisms – revolutionized molecular biology, allowing scientists to use them as markers for genetic expression, to locate molecules and observe activity within cells.
The recent discovery of photoswitchable fluorescent proteins – which can be manipulated with a laser – has been a significant development for cellular research.
"Photoswitchable fluorescent proteins have tremendous advantages over passive proteins," Remington said. "You can label all molecules but using a laser under a microscope, you can activate only a small group of them. That lets you follow the motion of subsets of molecules. We wanted to understand the process, so that we can permanently switch them off and on or vary the time delay."
However, he said, the mechanism of photoswitching was unknown, and in many cases the proteins returned to their stable state randomly and spontaneously.
Using a combination of rational mutagenesis and directed evolution, UO doctoral student J. Nathan Henderson determined high-resolution crystal structures of both the on and off states of a fluorescent protein isolated from a sea anemone.
In the stable or fluorescent state of the molecule, two side chains of atoms align in a coplanar fashion, flat and in orderly fashion. When hit with bright laser light, the researchers observed that the protein rapidly went dark as the rings rotated about 180 degrees and flip by some 45 degrees, coming to rest in a non-coplanar and unstable alignment. The two structures gave the researchers a chance to observe changes in the interactions between neighboring groups.
Remington said that in the dark state, the molecule absorbs ultraviolet light and doesn't emit any light at all. However, when the chromophore (a group of atoms and electrons forming part of an molecule) absorbs ultraviolet light, it occasionally ionizes and become negatively charged. This causes the rings to flip back into the fluorescent state.
Having control of light emission would allow for more precise studies within cells, he said.
Henderson studied the structures, noticing that in the dark state there was an unfavorable interaction where carbon and oxygen atoms were adjacent to each other. "Nathan looked at this and wondered what would happen if an oxygen atom was inserted at a precise place," Remington said. "That would make for a favorable interaction that stabilized the dark state. Based on the structure, Henderson made a single mutation that delays the switch-on time from five minutes to 65 hours.
Eventually, he added, the ability to control the on-off states could lead to improvements in optical memory, such as single molecule information storage, in addition to enhancing microscopic work and molecular labeling.
Source: University of Oregon