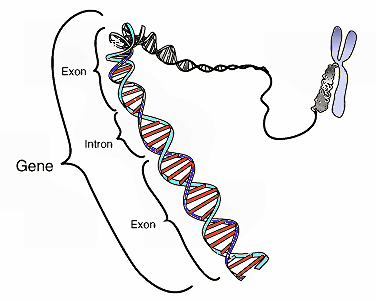
This image shows the coding region in a segment of eukaryotic DNA. Credit: National Human Genome Research Institute
(Phys.org)—A team of researchers with the University of California has learned more about the process that is involved during CRISPR/Cas9 gene splicing by using a crystallization technique to get a closer look. In their paper published in the journal Science, the team describes their study of the way three-stranded structures known as R-loops are formed during the technique and how they used a familiar technique to capture 3D images of the process and what they learned as a result.
Over the past three years, the CRISPR/Cas9 gene splicing and editing tool has been used in a variety of experiments to better understand how genes work and to possibly edit them to cure genetic diseases. The technique has been in the news again recently as two parties have claimed to be the original inventors of the technique and the U.S. Patent and Trademark Office has agreed to move forward with interference hearings to decide the case. It is one of those two teams, led by Jennifer Doudna, that has now made clearer the means by which R-loops are formed as part of the technique.
The CRISPR/Cas9 technique, as its name implies, is based on the Cas9 protein and they way it causes R-loops to form, but until now, no one had ever seen it actually do its work. In this new effort, the researchers used a technique that has been used to study proteins before—where a protein under study is caused to grow into a crystal where it is then exposed to x-rays—the diffraction patterns given off offer clues about its structure. In this case the technique was used to allow the researchers to capture 3D images of the Cas9 protein during the moments it was editing a DNA sequence—those images showed the interactions that occurred between the protein and DNA strands that led to the strands bending at a 30° angle, which then caused the DNA and RNA strands to move into a position that led to the formation of an R-loop.
Understanding how and why R-loops form during gene splicing is critical to improving the technique—sometimes mistakes are made, such as identifying and cutting the wrong strand of DNA. It might also help in identifying other proteins that might be used for the same purpose, but under different circumstances.
More information: F. Jiang et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage, Science (2016). DOI: 10.1126/science.aad8282
Abstract
Bacterial adaptive immunity and genome engineering involving the CRISPR (clustered regularly interspaced short palindromic repeats)–associated (Cas) protein Cas9 begin with RNA-guided DNA unwinding to form an RNA-DNA hybrid and a displaced DNA strand inside the protein. The role of this R-loop structure in positioning each DNA strand for cleavage by the two Cas9 nuclease domains is unknown. We determine molecular structures of the catalytically active Streptococcus pyogenes Cas9 R-loop that show the displaced DNA strand located near the RuvC nuclease domain active site. These protein-DNA interactions in turn position the HNH nuclease domain adjacent to the target DNA strand cleavage site in a conformation essential for concerted DNA cutting. Cas9 bends the DNA helix by 30°, providing the structural distortion needed for R-loop formation.
Journal information: Science
© 2016 Phys.org