Mechanical force controls the speed of protein synthesis
As cells create proteins, the proteins modulate synthesis speed by exerting a mechanical force on the molecular machine that makes them, according to a team of scientists who used a combination of computational and experimental techniques to understand this force.
Proteins power many of a cell's vital functions, from providing structure to delivering information to fighting viruses. Defective protein synthesis is linked to numerous diseases, including subtypes of hemophilia, lung carcinoma, and cervical and vulvar cancers.
"What has been observed in the past decade is that if you change the speed at which a protein gets synthesized, you can alter how the protein behaves," said Edward O'Brien, assistant professor of chemistry and an Institute for CyberScience co-hire, Penn State. "We tried to identify new factors that influence protein synthesis speed."
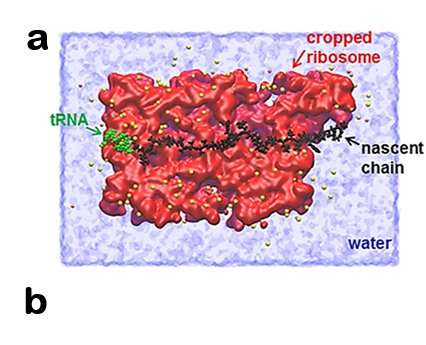
Ribosomes, tiny factories in the cell, stitch amino acids into a long chain to create proteins. During this process, newly synthesized protein segments pass through a narrow tunnel of the ribosome. When they exit the tunnel, proteins naturally pull away from the ribosome, O'Brien said.
"These protein molecules like to be in regions of space with a lot of free volume where they can move around, rather than in confined narrow spaces," said O'Brien.
The force that pulls the protein from the ribosome is an entropic pulling force that happens naturally, according to O'Brien. Entropic force in a system is a force resulting from the entire system's tendency to increase its entropy, rather than from a particular underlying microscopic force. Entropy is the tendency for systems to become more disordered over time.
"That pulling force gets propagated back to where the synthesis is occurring within the ribosome, and modulates that process," said O'Brien.
The researchers observed that unstructured protein segments generate piconewtons of force and that this force is transmitted across the ribosome molecular machine, and that it affects the speed at which amino acids are stitched together.
The team started their study from experimental measurements that detected how much proteins stretched on the ribosome. The researchers input this information into high-performance computer simulations that ran for months using both the Penn State Institute for CyberScience's Advanced CyberInfrastructure and the Extreme Science and Engineering Discovery Environment, an NSF-funded virtual organization. These simulations allowed them to see how protein synthesis was carried out under numerous conditions.
"By understanding the factors governing the speed of protein synthesis, we can now start to understand how protein synthesis affects downstream processes involving protein structure and function, including various diseases," said O'Brien.
The researchers published their findings in the Journal of the American Chemical Society.
Journal information: Journal of the American Chemical Society