This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
New method enables amplifiable protein identification from trace samples
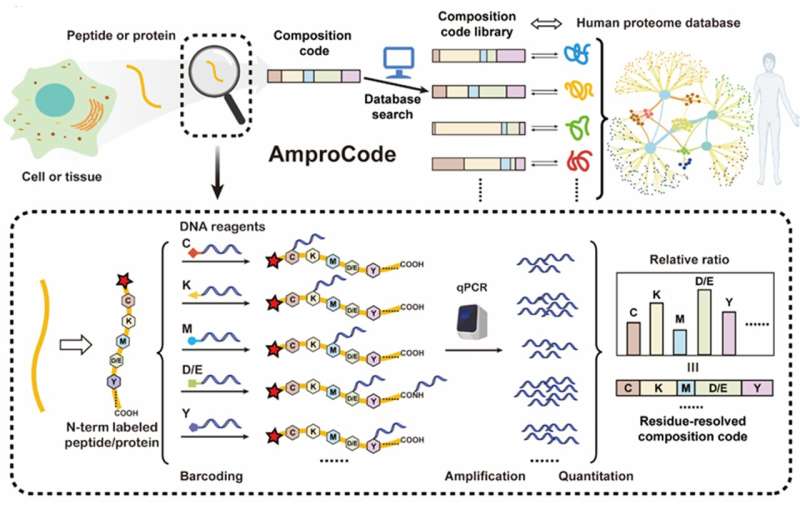
In a study published in the journal National Science Review, researchers developed an amplifiable protein identification method called "AmproCode."
Ultrasensitive protein identification is significant but extremely challenging, because unlike the nucleic acids that could rely on the polymerase chain reaction (PCR) for amplification, proteins cannot be directly amplified.
However, the authors believe that application of a collection of residue-specific reactions that can selectively label different types of amino acids with the amplifiable DNA barcodes, in conjunction with the innovative proteome database matching method, may ultimately produce a breakthrough technique for turning proteins into amplifiable molecules. They validated their idea both computationally and experimentally in this study.
They first designed computational assays for proteome database matching and for theoretical estimation of the identification rate of AmproCode. The computational results showed that 99% of the proteins in the whole proteome and 93% of the proteins in the secretome could be identified after quantifying only four residue types, providing the theoretical basis for their method.
They also rationally selected several residue types including Cys, Lys, Met, Asp/Glu and Tyr for AmproCode. They tested their residue-specific chemical reactions on both small molecules and peptides, and designed the DNA barcoding strategy.
Then, the researchers demonstrated AmproCode by the identification of different synthetic peptides including ELA, URP and Aβ found in the secretome. Their technique allows protein identification at an extremely low concentration (fmol/L), which is 10 to 10,000 times more sensitive than the common mass spectrometry and ELISA-based approaches. It shows that AmproCode has the capability for amplifiable protein identification from trace samples.
Moreover, they evaluated the performance of AmproCode in different conditions by computational estimations. They found that quantitation of more residue could bring significant improvement to AmproCode in the coverage, accuracy and adaptability.
Although this is only a proof-of-concept study, their novel concept of amplifiable protein fingerprinting provides a new framework towards development of the next generation protein sequencing, which may finally realize single-cell proteomics and may promote the discovery of clinical biomarkers.
The study was led by Prof. Peng R. Chen and Prof. Chu Wang (College of Chemistry and Molecular Engineering, Peking-Tsinghua Center for Life Sciences, Academy for Advanced Interdisciplinary Studies, Peking University).
More information: Weiming Guo et al, Amplifiable protein identification via residue-resolved barcoding and composition code counting, National Science Review (2024). DOI: 10.1093/nsr/nwae183
Provided by Science China Press