New chemoproteomic method enables global profiling of arginine dimethylation
Protein arginine methylation plays an important role in regulating protein functions in different cellular processes, and its dysregulation may lead to a variety of diseases.
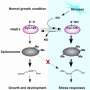
In recent years, mounting evidence has demonstrated that arginine methylation may play an important role in regulating liquid-liquid phase separation (LLPS) of the proteins involved in the dynamic (dis)assembly of different membraneless organelles (MLOs). However, the global identification and characterization of arginine methylation in regulating protein LLPS and MLO dynamic (dis)assembly are still unclear.
Recently, a research team led by Prof. Ye Mingliang from the Dalian Institute of Chemical Physics (DICP) of the Chinese Academy of Sciences (CAS), in collaboration with Prof. Liu Cong from the Shanghai Institute of Organic Chemistry (SIOC) of CAS, revealed the way arginine dimethylation regulates protein LLPS and MLOs by using a new chemoproteomic method.
This study was published in Proceedings of the National Academy of Sciences (PNAS) on October 18.
Arginine residue can be modified with a cis-diol group by reacting with vicinal dicarbonyl compound, which enables the enrichment of arginine-containing peptides by boronate-affinity chromatography.
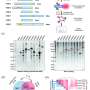
In this study, the researchers found that the modifications of some groups on arginine residue could severely influence this reaction. Inspired by this, they developed a steric effect-based chemical enrichment method (SECEM), which could enrich arginine dimethylated peptides from complex peptide mixture for proteomics analysis. They found this method could increase the identification performance of arginine demethylation (DMA) at the proteome level.
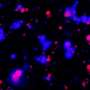
By using SECEM, the researchers revealed that, in mammalian cells, the DMA sites occurring in the RG/RGG motifs were enriched within the proteins identified in different MLOs, especially stress granules (SGs).
Moreover, further global profiling of the arginine DMA dynamic change upon SG formation by SECEM identified that the most dramatic change of arginine dimethylation occurred at multiple sites of RG/RGG-rich regions from several key SG-contained proteins, including G3BP1, FUS, hnRNPA1, and KHDRBS1.
Notably, in vitro arginine methylation and mutation of dimethylated arginine site impaired LLPS capability of these RG/RGG-rich regions, which further validated the important role of DMA in regulating protein LLPS.
More information: Qi Wang et al, Global profiling of arginine dimethylation in regulating protein phase separation by a steric effect–based chemical-enrichment method, Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2205255119
Journal information: Proceedings of the National Academy of Sciences
Provided by Chinese Academy of Sciences